Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein - PubMed (original) (raw)
Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein
Andrei Kuzmichev et al. Genes Dev. 2002.
Abstract
Enhancer of Zeste [E(z)] is a Polycomb-group transcriptional repressor and one of the founding members of the family of SET domain-containing proteins. Several SET-domain proteins possess intrinsic histone methyltransferase (HMT) activity. However, recombinant E(z) protein was found to be inactive in a HMT assay. Here we report the isolation of a multiprotein E(z) complex that contains extra sex combs, suppressor of zeste-12 [Su(z)12], and the histone binding proteins RbAp46/RbAp48. This complex, which we termed Polycomb repressive complex (PRC) 2, possesses HMT activity with specificity for Lys 9 (K9) and Lys 27 (K27) of histone H3. The HMT activity of PRC2 is dependent on an intact SET domain in the E(z) protein. We hypothesize that transcriptional repression by the E(z) protein involves methylation-dependent recruitment of PRC1. The presence of Su(z)12, a strong suppressor of position effect variegation, in PRC2 suggests that PRC2 may play a widespread role in heterochromatin-mediated silencing.
Figures
Figure 1
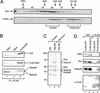
Isolation of the human Flag-ESC-containing complex. (A) Western blot analysis of the sephacryl-400 column fractions. Flag-ESC (F-ESC) was detected using anti-Flag antibodies. E(z) was detected using antibodies raised against the recombinant protein (a gift of Dr. Otte). E(z) and Flag-ESC are indicated with arrows. Vertical arrows indicate the elution peaks of molecular size standards. The “complex” and “monomeric” pools used for anti-Flag immunoffinity purification are indicated. (B) Western blot analysis of the immunoaffinity-purified Flag-ESC complex. Input indicates 1/100 (10 μL) of the complex pool from 293-TAG cell extracts used for anti-Flag immunopurification; Flag-ESC indicates one tenth (10 μL) of the peptide eluate from the anti-Flag affinity column; mock indicates one tenth of the peptide eluate from an anti-Flag affinity column that had been used to purify the complex pool from untagged cell extracts. IP, immunoprecipitation; α-Flag, anti-Flag antibodies. (C) Silver staining of an SDS-polyacrylamide gel containing anti-Flag column peptide eluates. Complex and monomeric pools were derived from 293 and TAG-293 nuclear extracts through a sephacryl-400 column. The bands specific to the Flag-ESC complex are indicated. Dots indicate contaminating bands, which are also present in immunoprecipitates from the untagged cell extracts (293). The positions of molecular size standards are also indicated. (D) Western blot of anti-Flag immunoprecipitates from 293 cells transiently transfected with Flag-GFP (negative control) or Flag-Su(z)12. Input indicates 1/100 (∼10 μg) of the nuclear extract used for immunoprecipitation; α-Flag lanes, one half of the peptide eluate fractions from the anti-Flag column.
Figure 2
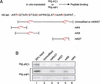
Conventional purification of the E(z)-ESC complex. (A) Schematic representation of the purification procedure utilized to isolate the PRC2 as well as other HMTs. The three H3-specific HMTs bound to the DEAE-cellulose column are indicated as I, II, and III. The brown curve shows HMT activity of DEAE-cellulose fractions on oligonucleosomes in the absence of histone H1, and the orange curve shows the activity in the presence of H1. Performing HMT assays with and without histone H1 allowed us to clearly differentiate the three different peaks of activity bound to the column. Each of the peaks were further fractionated on a gel filtration column as described by Nishioka et al. (2002a,b). The native molecular weight of the different H3-HMTs are indicated. The steps of purification of the 500-kD activity, PRC2, are shown and described in the Materials and Methods section. (B) HMT assay and Western blot analysis of the DEAE-5PW column. The pool derived from the gel filtration step was fractionated onto a DEAE-5PW column. Fractions of the column were assayed for HMT activity using histone octamers as substrates. The presence of different polypeptides was determined by Western blot. Two peaks of HMT activity were observed coeluting with E(z) and other PRC2 components. These peaks are referred to as “complex I” and “complex II”. The protein identified as HDAC, which copurifies with complex I, is marked by dots. (C) Analysis of the final step of PRC2 purification chromatography on a gel filtration Superose 12 column. The top panel shows silver staining of fractions (10 μL) derived from the Superose 12 column. The bands coeluting with HMT activity are indicated by asterisks; elution peaks of molecular size standards, on the top. The positions of SDS-PAGE molecular weight markers are indicated on the left. The middle panel shows the HMT assay with Superose 12 fractions (10 μL) using recombinant histone octamers as substrates. The position of histone H3 is indicated. The bottom panels show Western blot analysis of Superose 12 fractions using the indicated antibodies.
Figure 3
Characterization of the HMT activity of PRC2. (A) Substrate specificity assay with conventionally purified E(z)-ESC complex. The different substrates used are recombinant octamer (rOct), native HeLa octamer (nOct), oligonucleosome assembled in vitro using recombinant histones (rNuc), and oligonucleosome assembled in vitro using native histones (nNuc). Arrows indicate the addition of different amounts of PRC2. The top panel shows an autoradiograph and the bottom panel shows Coomassie blue staining of the HMT assay membrane. Positions of different histones are indicated. Migration of recombinant histones is indicated on the left side and migration of the native purified HeLa histones is indicated on the right side. (B) Edman degradation performed on isolated histone H3 using native core histones as a substrate. Products of the HMT reaction were separated with SDS-PAGE, and the labeled H3 polypeptide was excised and subjected to Edman degradation. The numbers at the bottom represent the cycles of degradation and correspond to the amino acid of the H3 tail. (C) Determination of methylation site preference of the PRC2. HMT assays were performed with octamers assembled with recombinant histones using either wild-type (Wt) histone H3, or histone H3 containing the following amino acid substitutions: K9 to alanine (K9A), K27 to alanine (K27A), and a substitution of K9 and K27 to alanine (K9/27A) as indicated. Arrows indicate the addition of different amounts of purified PRC2. The top panel is an autoradiograph, the middle panel is a Coomassie blue staining of the HMT assay membrane, and the bottom panel is quantification of the autoradiograph. (D) Determination of preferred methylation sites on histone H3 using GST-fusion proteins in which the candidates methylation sites were substituted as indicated. Anti-Flag affinity-purified E(z)-ESC complex was used in the analysis. GST-H3 indicates the wild-type H3 tail; GST-H3-K4, an H3 tail in which K9 and K27 have been mutated to arginines (K9R/K27R); GST-H3-K9, the K4R/K37R mutant; and GST-H3-K27, the K4R/K9R mutant. Top and bottom panels are the same as in A.
Figure 4
HMT activity of PRC2 containing mutant E(z) protein. (A) Alignment of the SET domains of Ezh2 [the mouse homolog of E(z)], SUV39H1, G9A, Set9, and PR-Set7. Alignment was performed using the CLUSTALW algorithm at the European Bioinformatics Institute server (
http://www2.ebi.ac.uk/clustalw
; Thompson et al. 1994). The NHS consensus within the SET domain is indicated by yellow. The conserved histidine 689 is shown in red and the C588 and R727 residues are indicated in yellow . The borders of the SET and Pre-SET domains are shown. (B) HMT assays with PRC2 purified from transfected cells containing the wild-type E(z) protein or the catalytic site mutant. The PRC2 were purified by anti-Flag immunoaffinity chromatography from 293 cells transfected with either an empty expression vector control (Flag), or expression vectors encoding wild-type E(z) [Flag-E(z)] or the H689A mutant [Flag-E(z) H689A]. Nuclear extracts (∼1 mg) were immunoprecipitated with anti-Flag M2 agarose, and the peptide eluates (10 μL out of 20 μL) were used for the HMT assays. The top panel shows an autoradiograph, the middle panel shows a Coomassie blue staining of the HMT assay membrane, and the bottom panel shows an anti-Flag Western blot of the HMT assay reaction. (C) HMT assays were performed as in B using affinity-purified PRC2 containing wild-type (Wt) E(z) protein or the C588Y and R727K mutants. The top panel is anti-Flag Western blot and the bottom panel is an autoradiograph of the HMT assay. (D) Methylation site preference of affinity purified PRC2 complexes containing the C588Y or R727K E(z) mutant. The GST-H3 tail fusion proteins—wild-type (wt), double K9/K27 mutant (K4), double K4/K27 mutant (K9), and double K4/K9 mutant (K27)—were methylated using increasing amounts of indicated mutant PRC2 complexes. The top panel shows the autoradiograph of the HMT assay, the middle panel shows the Coomassie stain showing equal amounts of GST substrates used in the analysis, and the bottom panel shows anti-FLAG Western blot demonstrating that each set of assay received approximately equal amounts of complex.
Figure 5
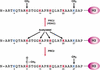
Interaction of PC1 protein with H3 tail methylated at K9 and K27. (A) Schematic representation of the different peptides used in the pull-down assays shown in panel B. In vitro translated mPC1 (35S-mPC1) or HP1 (35S-HP1) proteins were incubated with immobilized H3 tail peptides containing the following modifications: trimethyl-K4 (mK4), trimethyl-K9 (mK9), trimethyl-K27 (mK27), and trimethyl-K9 together with trimethyl-K27 (mK9/27) or with unmodified H3 tail peptide (unmodified). (B) The binding assays were washed as described in Materials and Methods, and the proteins remaining bound to the peptides were analyzed by SDS-PAGE followed by autoradiography. The top panel shows PC1-binding to indicated peptides without competitor BSA and the middle and bottom panels show HP1 binding to indicated peptides in absence (middle) or presence (bottom) of competitor BSA. The sequence of the peptides and the scheme of the experiment are illustrated on the top of panel A.
Figure 6
Model depicting the establishment of two different “marks” on the histone H3-tail. (Middle) The H3-tail and the lysines (red) that are functionally known to be acetylated in vivo, as well as K4 and K27 (blue) that are methylated but apparently not acetylated in higher eukaryotes. (Top) The marks established by PRC2 when it functions together with histone deacetylase. In this case, K9 (and possible K14, Lys 18, and Lys 23) is deacetylated, creating a site (K9) that can be accessed by PRC2. In this manner PRC2 methylates both K9 and K27. (Bottom) PRC2 in the absence of a histone deacetylase can methylate only K27, creating a “mark” on the histone H3 tail that is functionally different from that shown in the top panel. In this case, K9 remains acetylated.
Similar articles
- Histone methyltransferase activity of a Drosophila Polycomb group repressor complex.
Müller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, Miller EL, O'Connor MB, Kingston RE, Simon JA. Müller J, et al. Cell. 2002 Oct 18;111(2):197-208. doi: 10.1016/s0092-8674(02)00976-5. Cell. 2002. PMID: 12408864 - Role of histone H3 lysine 27 methylation in Polycomb-group silencing.
Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Cao R, et al. Science. 2002 Nov 1;298(5595):1039-43. doi: 10.1126/science.1076997. Epub 2002 Sep 26. Science. 2002. PMID: 12351676 - Polycomb group suppressor of zeste 12 links heterochromatin protein 1alpha and enhancer of zeste 2.
Yamamoto K, Sonoda M, Inokuchi J, Shirasawa S, Sasazuki T. Yamamoto K, et al. J Biol Chem. 2004 Jan 2;279(1):401-6. doi: 10.1074/jbc.M307344200. Epub 2003 Oct 21. J Biol Chem. 2004. PMID: 14570930 - The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3.
Cao R, Zhang Y. Cao R, et al. Curr Opin Genet Dev. 2004 Apr;14(2):155-64. doi: 10.1016/j.gde.2004.02.001. Curr Opin Genet Dev. 2004. PMID: 15196462 Review. - Inner workings and regulatory inputs that control Polycomb repressive complex 2.
O'Meara MM, Simon JA. O'Meara MM, et al. Chromosoma. 2012 Jun;121(3):221-34. doi: 10.1007/s00412-012-0361-1. Epub 2012 Feb 19. Chromosoma. 2012. PMID: 22349693 Free PMC article. Review.
Cited by
- Epigenetic regulation by polycomb repressive complex 1 promotes cerebral cavernous malformations.
Pham VC, Rödel CJ, Valentino M, Malinverno M, Paolini A, Münch J, Pasquier C, Onyeogaziri FC, Lazovic B, Girard R, Koskimäki J, Hußmann M, Keith B, Jachimowicz D, Kohl F, Hagelkruys A, Penninger JM, Schulte-Merker S, Awad IA, Hicks R, Magnusson PU, Faurobert E, Pagani M, Abdelilah-Seyfried S. Pham VC, et al. EMBO Mol Med. 2024 Oct 14. doi: 10.1038/s44321-024-00152-9. Online ahead of print. EMBO Mol Med. 2024. PMID: 39402138 - Replication-coupled inheritance of chromatin states.
Song A, Wang Y, Liu C, Yu J, Zhang Z, Lan L, Lin H, Zhao J, Li G. Song A, et al. Cell Insight. 2024 Aug 23;3(6):100195. doi: 10.1016/j.cellin.2024.100195. eCollection 2024 Dec. Cell Insight. 2024. PMID: 39391004 Free PMC article. Review. - Intracellular checkpoints for NK cell cancer immunotherapy.
Huang Y, Tian Z, Bi J. Huang Y, et al. Front Med. 2024 Oct;18(5):763-777. doi: 10.1007/s11684-024-1090-6. Epub 2024 Sep 28. Front Med. 2024. PMID: 39340588 Review. - Cohesin mutations in acute myeloid leukemia.
Boucher A, Murray J, Rao S. Boucher A, et al. Leukemia. 2024 Nov;38(11):2318-2328. doi: 10.1038/s41375-024-02406-4. Epub 2024 Sep 9. Leukemia. 2024. PMID: 39251741 Review. - Regulation of chromatin architecture by protein binding: insights from molecular modeling.
Portillo-Ledesma S, Schlick T. Portillo-Ledesma S, et al. Biophys Rev. 2024 May 9;16(3):331-343. doi: 10.1007/s12551-024-01195-5. eCollection 2024 Jun. Biophys Rev. 2024. PMID: 39099845 Review.
References
- Beuchle D, Struhl G, Muller J. Polycomb group proteins and heritable silencing of Drosophila Hox genes. Development. 2001;128:993–1004. - PubMed
- Birve A, Sengupta AK, Beuchle D, Larsson J, Kennison JA, Rasmuson-Lestander A, Muller J. Su(z)12, a novel Drosophila Polycomb group gene that is conserved in vertebrates and plants. Development. 2001;128:3371–3379. - PubMed
- Carrington EA, Jones RS. The Drosophila Enhancer of zeste gene encodes a chromosomal protein: examination of wild-type and mutant protein distribution. Development. 1996;122:4073–4083. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- GM-37120/GM/NIGMS NIH HHS/United States
- P30 CA08748/CA/NCI NIH HHS/United States
- R01 GM037120/GM/NIGMS NIH HHS/United States
- P30 CA008748/CA/NCI NIH HHS/United States
- R37 GM037120/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous