In vitro studies with recombinant Plasmodium falciparum apical membrane antigen 1 (AMA1): production and activity of an AMA1 vaccine and generation of a multiallelic response - PubMed (original) (raw)
In vitro studies with recombinant Plasmodium falciparum apical membrane antigen 1 (AMA1): production and activity of an AMA1 vaccine and generation of a multiallelic response
Michael C Kennedy et al. Infect Immun. 2002 Dec.
Abstract
Apical membrane antigen 1 (AMA1) is regarded as a leading malaria blood-stage vaccine candidate. While the overall structure of AMA1 is conserved in Plasmodium spp., numerous AMA1 allelic variants of P. falciparum have been described. The effect of AMA1 allelic diversity on the ability of a recombinant AMA1 vaccine to protect against human infection by different P. falciparum strains is unknown. We characterize two allelic forms of AMA1 that were both produced in Pichia pastoris at a sufficient economy of scale to be usable for clinical vaccine studies. Both proteins were used to immunize rabbits, singly and in combination, in order to evaluate their immunogenicity and the ability of elicited antibodies to block the growth of different P. falciparum clones. Both antigens, when used alone, elicited high homologous anti-AMA1 titers, with reduced strain cross-reactivity. Similarly, sera from rabbits immunized with a single antigen were capable of blocking the growth of homologous parasite strains at levels theoretically sufficient to clear parasite infections. However, heterologous inhibition was significantly reduced, providing experimental evidence that AMA1 allelic diversity is a result of immune pressure. Encouragingly, rabbits immunized with a combination of both antigens exhibited titers and levels of parasite inhibition as good as those of the single-antigen-immunized rabbits for each of the homologous parasite lines, and consequently exhibited a broadening of allelic diversity coverage.
Figures
FIG. 1.
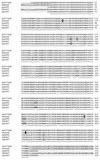
Alignment of native and synthetic AMA1 amino acid sequences. The GenBank sequences for P. falciparum FVO AMA1 (AJ277646 and U84348) and P. falciparum F3D7 AMA1 (U65407) are aligned with the sequences used for recombinant protein production in this study (SynFVO and Syn3D7). The three mutations unique to U84348 among all P. falciparum AMA1's, and so not used in the synthetic FVO sequence, are indicated by a solid background. The single glycosylation site mutated on the basis of alignment with allelic variations in other P. falciparum AMA1 sequences is shown as a boxed “K.” Another amino acid in the SynFVO sequence was mutated to T (boxed), because a T always accompanies a K in the previous position (both in other P. falciparum sequences and in the AMA1 sequences of other Plasmodium species). Mutations indicated by shading were chosen by alignment with sequences of other Plasmodium species, because all P. falciparum AMA1 sequences were conserved in this region. The first three of these were from other primate malaria species (Plasmodium vivax, Plasmodium cynomolgi, and/or Plasmodium fragile), and the last was from P. chabaudi. Lowercase letters in the synthetic recombinant sequences represent vector-derived sequences. The signal peptide, transmembrane, and cytoplasmic domains present in the native sequences, but not used in the recombinant proteins, are underlined.
FIG. 2.
Purity and integrity of PpAMA1's. SDS-PAGE on 4-to-20% gradient Tris-glycine polyacrylamide gels was performed on serial fourfold dilutions of purified PpAMA1 FVO (A and B) and PpAMA1 3D7 (C and D). Quantities loaded were, from left to right, 12.5, 3.1, 0.8, and 0.2 μg. Electrophoresis was performed either under nonreducing conditions (A and C) or with 5% (vol/vol) β-mercaptoethanol (B and D). Scanning laser densitometry showed yields of 96.9% (nonreduced) or 81.9% (reduced) full-length PpAMA1 FVO and 97.6% (nonreduced) or 48.4% (reduced) full-length PpAMA1 3D7. Single asterisks indicate bands whose N-terminal sequence was identified by Edman degradation as Y1VQNYWEHPYQKSDVYHPIN (SynFVO) or Y1VQNYWEHPYQNSDVYRPIN (Syn3D7); double asterisks indicate bands whose N-terminal sequence was S355AFLPTGAFKADRYKSH.
FIG. 3.
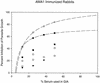
Mean growth inhibition levels of rabbit sera raised against recombinant PpAMA1's. Groups of five rabbits each were immunized with 50 μg of either PpAMA1 FVO (solid symbols) or PpAMA1 3D7 (open symbols). Each rabbit serum was assayed individually for growth inhibition against three parasite lines (FVO [circles], 3D7 [inverted triangles], or HB3 [squares]), and the mean percent inhibition of invasion of RBCs for each group of five rabbits is shown along the y axis. GIAs for each serum were conducted at three (with HB3 parasites) or four concentrations, as shown on the x axis. Lines represent regression of homologous results (i.e., PpAMA FVO versus FVO parasites and PpAMA 3D7 versus 3D7 parasites) by use of a hyperbolic equation.
FIG. 4.
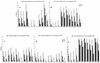
Homologous and heterologous growth inhibition of P. falciparum by sera raised against two alleles of AMA1. Individual sera from each of four rabbits in each of the three immunization groups (PpAMA1 FVO, PpAMA 3D7, or a combination of both) were tested at three different serum concentrations (25, 12.5, and 6.25%) for their abilities to inhibit the growth of five different strains of P. falciparum.
FIG. 5.
Homologous and heterologous GIAs comparing rabbit groups receiving single versus combined immunogens. Rabbits from each of the three immunization groups were compared at the highest serum concentration (25%) for their abilities to inhibit the growth of five different P. falciparum strains. Horizontal lines show the mean value for each group. Groups were compared for statistical significance by an unpaired t test.
FIG. 6.
Effects of antigenic differences on growth-inhibitory activity. Rabbits were immunized with either PpAMA1 FVO (blue), PpAMA1 3D7 (red), or a combination of both (green). Sera from immunized rabbits were used in GIAs against the M24, HB3, D10, FVO, and 3D7 P. falciparum clones. The number of amino acid differences between an immunizing AMA1 allele (FVO, 3D7, or a combination of both) and the parasite clone tested for inhibition is plotted on the x axis, and the mean level of inhibition obtained against that parasite is plotted on the y axis. For the combined immunization, only those amino acids present in neither immunizing allele are taken into account. The dotted line is the result of a linear regression of all data points.
Similar articles
- Overcoming allelic specificity by immunization with five allelic forms of Plasmodium falciparum apical membrane antigen 1.
Miura K, Herrera R, Diouf A, Zhou H, Mu J, Hu Z, MacDonald NJ, Reiter K, Nguyen V, Shimp RL Jr, Singh K, Narum DL, Long CA, Miller LH. Miura K, et al. Infect Immun. 2013 May;81(5):1491-501. doi: 10.1128/IAI.01414-12. Epub 2013 Feb 19. Infect Immun. 2013. PMID: 23429537 Free PMC article. - Limited antigenic diversity of Plasmodium falciparum apical membrane antigen 1 supports the development of effective multi-allele vaccines.
Terheggen U, Drew DR, Hodder AN, Cross NJ, Mugyenyi CK, Barry AE, Anders RF, Dutta S, Osier FH, Elliott SR, Senn N, Stanisic DI, Marsh K, Siba PM, Mueller I, Richards JS, Beeson JG. Terheggen U, et al. BMC Med. 2014 Oct 16;12:183. doi: 10.1186/s12916-014-0183-5. BMC Med. 2014. PMID: 25319190 Free PMC article. - A diversity-covering approach to immunization with Plasmodium falciparum apical membrane antigen 1 induces broader allelic recognition and growth inhibition responses in rabbits.
Remarque EJ, Faber BW, Kocken CH, Thomas AW. Remarque EJ, et al. Infect Immun. 2008 Jun;76(6):2660-70. doi: 10.1128/IAI.00170-08. Epub 2008 Mar 31. Infect Immun. 2008. PMID: 18378635 Free PMC article. - Recombinant protein vaccines against the asexual blood stages of Plasmodium falciparum.
Anders RF, Adda CG, Foley M, Norton RS. Anders RF, et al. Hum Vaccin. 2010 Jan;6(1):39-53. doi: 10.4161/hv.6.1.10712. Epub 2010 Jan 19. Hum Vaccin. 2010. PMID: 20061790 Review. - Apical membrane antigen 1: a malaria vaccine candidate in review.
Remarque EJ, Faber BW, Kocken CH, Thomas AW. Remarque EJ, et al. Trends Parasitol. 2008 Feb;24(2):74-84. doi: 10.1016/j.pt.2007.12.002. Epub 2008 Jan 15. Trends Parasitol. 2008. PMID: 18226584 Review.
Cited by
- Antigenic characterization of an intrinsically unstructured protein, Plasmodium falciparum merozoite surface protein 2.
Adda CG, MacRaild CA, Reiling L, Wycherley K, Boyle MJ, Kienzle V, Masendycz P, Foley M, Beeson JG, Norton RS, Anders RF. Adda CG, et al. Infect Immun. 2012 Dec;80(12):4177-85. doi: 10.1128/IAI.00665-12. Epub 2012 Sep 10. Infect Immun. 2012. PMID: 22966050 Free PMC article. - Relationship between malaria incidence and IgG levels to Plasmodium falciparum merozoite antigens in Malian children: impact of hemoglobins S and C.
Miura K, Diakite M, Diouf A, Doumbia S, Konate D, Keita AS, Moretz SE, Tullo G, Zhou H, Lopera-Mesa TM, Anderson JM, Fairhurst RM, Long CA. Miura K, et al. PLoS One. 2013;8(3):e60182. doi: 10.1371/journal.pone.0060182. Epub 2013 Mar 28. PLoS One. 2013. PMID: 23555917 Free PMC article. - Production, Quality Control, Stability and Pharmacotoxicity of a Malaria Vaccine Comprising Three Highly Similar PfAMA1 Protein Molecules to Overcome Antigenic Variation.
Faber BW, Hellwig S, Houard S, Havelange N, Drossard J, Mertens H, Croon A, Kastilan R, Byrne R, van der Werff N, van der Eijk M, Thomas AW, Kocken CH, Remarque EJ. Faber BW, et al. PLoS One. 2016 Oct 3;11(10):e0164053. doi: 10.1371/journal.pone.0164053. eCollection 2016. PLoS One. 2016. PMID: 27695087 Free PMC article. - Influenza virus-like particle vaccine containing both apical membrane antigen 1 and microneme-associated antigen proteins of Plasmodium berghei confers protection in mice.
Kim MJ, Chu KB, Kang HJ, Yoon KW, Lee DH, Lee SH, Moon EK, Quan FS. Kim MJ, et al. BMC Immunol. 2022 Apr 25;23(1):21. doi: 10.1186/s12865-022-00494-4. BMC Immunol. 2022. PMID: 35468726 Free PMC article. - The Cellular and Molecular Interaction Between Erythrocytes and Plasmodium falciparum Merozoites.
Molina-Franky J, Patarroyo ME, Kalkum M, Patarroyo MA. Molina-Franky J, et al. Front Cell Infect Microbiol. 2022 Mar 31;12:816574. doi: 10.3389/fcimb.2022.816574. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35433504 Free PMC article. Review.
References
- Amante, F. H., P. E. Crewther, R. F. Anders, and M. F. Good. 1997. A cryptic T cell epitope on the apical membrane antigen 1 of Plasmodium chabaudi adami can prime for an anamnestic antibody response: implications for malaria vaccine design. J. Immunol. 159:5535-5544. - PubMed
- Anders, R. F., P. E. Crewther, S. Edwards, M. Margetts, M. L. Matthew, B. Pollock, and D. Pye. 1998. Immunisation with recombinant AMA-1 protects mice against infection with Plasmodium chabaudi. Vaccine 16:240-247. - PubMed
- Basco, L. K., F. Marquet, M. M. Makler, and J. Le Bras. 1995. Plasmodium falciparum and Plasmodium vivax: lactate dehydrogenase activity and its application for in vitro drug susceptibility assay. Exp. Parasitol. 80:260-271. - PubMed
- Chen, G. X., C. Mueller, M. Wendlinger, and J. W. Zolg. 1987. Kinetic and molecular properties of the dihydrofolate reductase from pyrimethamine-sensitive and pyrimethamine-resistant clones of the human malaria parasite Plasmodium falciparum. Mol. Pharmacol. 31:430-437. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous