hNuf2 inhibition blocks stable kinetochore-microtubule attachment and induces mitotic cell death in HeLa cells - PubMed (original) (raw)
hNuf2 inhibition blocks stable kinetochore-microtubule attachment and induces mitotic cell death in HeLa cells
Jennifer G DeLuca et al. J Cell Biol. 2002.
Abstract
Identification of proteins that couple kinetochores to spindle microtubules is critical for understanding how accurate chromosome segregation is achieved in mitosis. Here we show that the protein hNuf2 specifically functions at kinetochores for stable microtubule attachment in HeLa cells. When hNuf2 is depleted by RNA interference, spindle formation occurs normally as cells enter mitosis, but kinetochores fail to form their attachments to spindle microtubules and cells block in prometaphase with an active spindle checkpoint. Kinetochores depleted of hNuf2 retain the microtubule motors CENP-E and cytoplasmic dynein, proteins previously implicated in recruiting kinetochore microtubules. Kinetochores also retain detectable levels of the spindle checkpoint proteins Mad2 and BubR1, as expected for activation of the spindle checkpoint by unattached kinetochores. In addition, the cell cycle block produced by hNuf2 depletion induces mitotic cells to undergo cell death. These data highlight a specific role for hNuf2 in kinetochore-microtubule attachment and suggest that hNuf2 is part of a molecular linker between the kinetochore attachment site and tubulin subunits within the lattice of attached plus ends.
Figures
Figure 1.
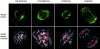
hNuf2 localizes to kinetochores throughout mitosis. HeLa cells were fixed and double stained with antibodies to hNuf2 and α-tubulin and observed using a spinning disc confocal fluorescence microscope.
Figure 2.
hNuf2-depleted cells exhibit a prolonged mitotic block and undergo cell death. (A) Reduction of hNuf2 levels after siRNA transfection. HeLa cells were transfected with hNuf2 siRNA, harvested 24, 48, and 72 h after transfection, and subjected to protein immunoblot analysis with hNuf2 antibodies and actin antibodies to control for gel loading. Time points are indicated above each lane, and time 0 indicates untransfected cells. (B) hNuf2 siRNA transfection results in a mitotic defect. Cells were transfected with either hNuf2 siRNA or a control 21-nucleotide siRNA duplex, and imaged 24, 48, and 72 h after transfection using a 10× phase objective. (C) Quantitation of control and hNuf2 siRNA–transfected cell phenotypes. Cells adherent to the culture dish with an intact nuclear envelope and decondensed chromatin were scored as interphase. Rounded cells with condensed chromosomes and smooth, uniform membranes were scored as mitotic. Cells that were multilobed, dense, and exhibited a nonuniform membrane were scored as dead (Mills et al., 1999; Zhang and Xu, 2002) (n = 6,000 cells per time point averaged from three independent experiments). (D) Mitotic progression of control siRNA– versus hNuf2 siRNA–transfected cells. 48 h after transfection, cells were transferred to live-cell chambers and time lapsed using a 40× phase objective. Top row, typical control siRNA–transfected cell; bottom row, typical hNuf2 siRNA–transfected cell. Time shown in minutes. (E) Time course of change in cellular DNA content after hNuf2 siRNA transfection. Time after transfection along the x axis is in hours, and untransfected cells were used as a control. Cells were analyzed by flow cytometry as described in the Materials and methods. (F) Cellular DNA content as analyzed by flow cytometry of cells treated for 48 h with 10 μM vinblastine. (G) Uptake of Trypan blue in hNuf2 siRNA–transfected cells. Mock-transfected and hNuf2 siRNA–transfected cells were incubated with Trypan blue 72 h after transfection and observed by phase-contrast and epifluorescence microscopy. (H) Nuclear fragmentation of hNuf2-depleted cells. Cells transfected with hNuf2 siRNA were fixed and stained with DAPI 48 h after transfection.
Figure 3.
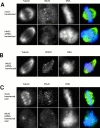
hNuf2-depleted cells lack stable kinetochore microtubules. (A and B) Immunofluorescent cell images of hNuf2 siRNA–transfected and mock-transfected cells. 48 h after transfection, cells were fixed for immunofluorescence and stained with the indicated antibodies. DAPI was used to visualize the DNA. (C) hNuf2 siRNA–transfected cells lack cold-stable kinetochore microtubules. Cells were incubated on ice for 10 min to induce complete microtubule disassembly of all nonkinetochore microtubules, and were subsequently fixed and processed for immunofluorescence using hNuf2 and tubulin antibodies and DAPI to stain DNA.
Figure 4.
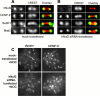
hNuf2-depleted kinetochores retain the spindle checkpoint proteins Mad2 and BubR1 and the motor proteins CENP-E and cytoplasmic dynein. Cells were mock transfected (A) or transfected with an hNuf2 siRNA (B) and fixed for immunofluorescence 48 h after transfection. The cells were stained using the indicated antibodies (left columns) and a kinetochore marker (CREST serum; center columns). Each image shows one kinetochore pair. (C) Kinetochore localization of CENP-E and dynein in nocodazole-treated cells. Cells were subjected to nocodazole treatment and immunofluorescence analysis.
Similar articles
- Microtubule-dependent changes in assembly of microtubule motor proteins and mitotic spindle checkpoint proteins at PtK1 kinetochores.
Hoffman DB, Pearson CG, Yen TJ, Howell BJ, Salmon ED. Hoffman DB, et al. Mol Biol Cell. 2001 Jul;12(7):1995-2009. doi: 10.1091/mbc.12.7.1995. Mol Biol Cell. 2001. PMID: 11451998 Free PMC article. - Inhibition of protein kinase C zeta blocks the attachment of stable microtubules to kinetochores leading to abnormal chromosome alignment.
Liu XF, Xie X, Miki T. Liu XF, et al. Cell Signal. 2006 Dec;18(12):2314-23. doi: 10.1016/j.cellsig.2006.05.017. Epub 2006 Jul 3. Cell Signal. 2006. PMID: 16820280 - Human CENP-I specifies localization of CENP-F, MAD1 and MAD2 to kinetochores and is essential for mitosis.
Liu ST, Hittle JC, Jablonski SA, Campbell MS, Yoda K, Yen TJ. Liu ST, et al. Nat Cell Biol. 2003 Apr;5(4):341-5. doi: 10.1038/ncb953. Nat Cell Biol. 2003. PMID: 12640463 - How the SAC gets the axe: Integrating kinetochore microtubule attachments with spindle assembly checkpoint signaling.
Agarwal S, Varma D. Agarwal S, et al. Bioarchitecture. 2015;5(1-2):1-12. doi: 10.1080/19490992.2015.1090669. Epub 2015 Oct 2. Bioarchitecture. 2015. PMID: 26430805 Free PMC article. Review. - Merotelic kinetochores in mammalian tissue cells.
Salmon ED, Cimini D, Cameron LA, DeLuca JG. Salmon ED, et al. Philos Trans R Soc Lond B Biol Sci. 2005 Mar 29;360(1455):553-68. doi: 10.1098/rstb.2004.1610. Philos Trans R Soc Lond B Biol Sci. 2005. PMID: 15897180 Free PMC article. Review.
Cited by
- Different Plk1 functions show distinct dependencies on Polo-Box domain-mediated targeting.
Hanisch A, Wehner A, Nigg EA, Silljé HH. Hanisch A, et al. Mol Biol Cell. 2006 Jan;17(1):448-59. doi: 10.1091/mbc.e05-08-0801. Epub 2005 Nov 2. Mol Biol Cell. 2006. PMID: 16267267 Free PMC article. - VTT-006, an anti-mitotic compound, binds to the Ndc80 complex and suppresses cancer cell growth in vitro.
Laine LJ, Mäki-Jouppila JHE, Kutvonen E, Tiikkainen P, Nyholm TKM, Tien JF, Umbreit NT, Härmä V, Kallio L, Davis TN, Asbury CL, Poso A, Gorbsky GJ, Kallio MJ. Laine LJ, et al. Oncoscience. 2021 Dec 10;8:134-153. doi: 10.18632/oncoscience.549. eCollection 2021. Oncoscience. 2021. PMID: 34926718 Free PMC article. - NUF2 Drives Clear Cell Renal Cell Carcinoma by Activating HMGA2 Transcription through KDM2A-mediated H3K36me2 Demethylation.
Lin J, Chen X, Yu H, Min S, Chen Y, Li Z, Xie X. Lin J, et al. Int J Biol Sci. 2022 May 16;18(9):3621-3635. doi: 10.7150/ijbs.70972. eCollection 2022. Int J Biol Sci. 2022. PMID: 35813477 Free PMC article. - Aurora-A inactivation causes mitotic spindle pole fragmentation by unbalancing microtubule-generated forces.
Asteriti IA, Giubettini M, Lavia P, Guarguaglini G. Asteriti IA, et al. Mol Cancer. 2011 Oct 19;10:131. doi: 10.1186/1476-4598-10-131. Mol Cancer. 2011. PMID: 22011530 Free PMC article. - Protein phosphatase 6 regulates mitotic spindle formation by controlling the T-loop phosphorylation state of Aurora A bound to its activator TPX2.
Zeng K, Bastos RN, Barr FA, Gruneberg U. Zeng K, et al. J Cell Biol. 2010 Dec 27;191(7):1315-32. doi: 10.1083/jcb.201008106. J Cell Biol. 2010. PMID: 21187329 Free PMC article.
References
- Elbashir, S.M., J. Harborth, W. Lendeckel, A. Yalcin, K. Weber, and T. Tuschl. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 411:494–498. - PubMed
- He, X., D.R. Rines, C.W. Espelin, and P.K. Sorger. 2001. Molecular analysis of kinetochore-microtubule attachment in budding yeast. Cell. 106:195–206. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R37 GM024364/GM/NIGMS NIH HHS/United States
- R01 GM024364/GM/NIGMS NIH HHS/United States
- GM24364/GM/NIGMS NIH HHS/United States
- GM66588/GM/NIGMS NIH HHS/United States
- F32 GM066588/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases