An N-terminal domain of Dbf4p mediates interaction with both origin recognition complex (ORC) and Rad53p and can deregulate late origin firing - PubMed (original) (raw)
An N-terminal domain of Dbf4p mediates interaction with both origin recognition complex (ORC) and Rad53p and can deregulate late origin firing
Bernard P Duncker et al. Proc Natl Acad Sci U S A. 2002.
Abstract
The Dbf4Cdc7 kinase acts at the level of individual origins to promote the initiation of DNA replication. We demonstrate through both immunoprecipitation and two-hybrid assays that a domain comprising the first 296 aa of Dbf4p interacts with Orc2p and Orc3p subunits of the origin recognition complex (ORC). Given that the activation of Rad53 kinase in response to the DNA replication checkpoint leads to the release of Dbf4p from an ORC-containing chromatin fraction, we also examined interaction between Dbf4p and Rad53p. This same domain of Dbf4p binds specifically to the forkhead homology-associated (FHA) domains of Rad53p. Cell cycle arrest in G(2)M, provoked by the overexpression of the Dbf4 domain, is suppressed in a rad53 mutant. Moreover, its overexpression perturbs the regulation of late, but not early, origin firing in wild-type cells after treatment with hydroxyurea.
Figures
Fig 1.
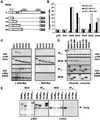
Dbf4p interacts with multiple ORC subunits through its N-terminal 296-aa domain. (A) Shown are constructs of full-length DBF4 or the N-terminal 109, 178, 248, or 296 residues fused to the C-terminal 46 residues and a Myc tag. The two regions that bind Cdc7p (27) and that mediate origin consensus interaction (ACS target; ref. 16) are indicated. The asterisk denotes a putative nuclear localization signal (P55KKRSLE). (B) Two-hybrid assays are described in Methods, using bait constructs pEG-Dbf4-FL, pEG-Dbf4-296, and empty pEG-202. pJG-ORC1 through -ORC6 were used for prey, with empty pJG4-6 (pJG) as control. For all two-hybrid assays, SD among independent colonies was <10%. (C) The pJG plasmids express the ORC subunits fused to an N-terminal hemagglutinin tag. Yeast strain GA-1020 was cotransformed with one of the pJG-ORC clones and pCM190 either with Dbf4-FL (+Dbf4-Myc) or without (−Dbf4-Myc). Expression was induced from both promoters, and IP from WCEs with anti-Myc is described in Methods. Shown are immunoblots of IPs and of input WCEs detected with either monoclonal anti-HA (for ORC detection) or anti-Myc antibodies (for Dbf4p detection). In each case, 1/20 of the input WCE and 1/4 of the final bead resuspension was blotted. (D) IP assays were carried as described in C, with GA-1020 cells cotransformed with pJG-ORC2 or -ORC3 and pCM190-Dbf4-FL, -178, -296, or empty pCM190. Arrowheads indicate Orc2p or Orc3p bands; dots indicate the IgG heavy chain (55 kDa). (E) IP was carried out by using WCEs from strains carrying Myc-tagged genomic copies of DBF4 (GA-850), HDF1 (Ku80, GA-1009), and DNA2 (GA-1099), and a nontagged strain (GA-1020), as in C. Blots were reacted with anti-Myc IgG or polyclonal rabbit anti-Orc2p serum
Fig 2.
Dbf4p interacts with the FHA1 and FHA2, but not the kinase domain of Rad53p. (A) Scheme of Dbf4 deletions and Rad53p fragments used in plasmid constructs for two hybrid and co-IP experiments. Fusions include amino acid 1–296 of Dbf4p, full-length Dbf4 (FL), and FL lacking amino acid 110 to 296. The Rad53p FHA1, kinase (KIN), and FHA2 domains are shown, including the R70A and R605A point mutations in FHA1 and FHA2 (33). (B) Two-hybrid assays were carried out as in Fig. 1_B_. Bait vectors and prey vectors are labeled as in A. Interactions with pJG-Dbf4-248 showed similar results as Dbf4-296, whereas pJG-Dbf4-274-704 bound none of the Rad53p domains (data not shown). (C) Two-hybrid assays were carried out as above, by using pEG-DBF4-FL and pEG-DBF4-Δ110–296 as baits. The scales at the left and right pertain to values obtained by using pJG-Orc2 and pJG-FHA1 as prey, respectively. (D) IP experiments were carried out as in Fig. 1_C_. WCEs were prepared from GA-1020 cells cotransformed with the indicated plasmids, as well as either pGAL-LexA-FHA1, -FHA2, or -KIN, as indicated. Immunoblots of the IP were probed with α-LexA antibody (CLONTECH). Arrows from top to bottom indicate Rad53 kinase, FHA2, and FHA1 LexA fusion proteins, whereas spots indicate the heavy and light IgG chains. (E) Two-hybrid analysis was performed as above, by using pEG-DBF4-FL as bait in each instance, along with the vector pJG4-6 expressing either ORC2 or FHA1. After 2 h of a 6-h galactose-induction, cultures were split, and 50 mM HU was added to one half of each.
Fig 3.
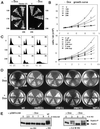
Overexpression of the ARS-interacting domain of Dbf4p in WT cells slows down the cell cycle. (A) Induction of Dbf4p fragments from pCM190-derived plasmids as described in Fig. 1. Expression of Dbf4-248 and -296 in GA-850 cells, but not of -109 and -178, arrests cell growth on plates without uracil and Dox. (B) Growth curves of GA-1020 cells expressing the Dbf4p fragments. (Upper) Cells expressing the indicated Dbf4 fragment were grown to saturation in Dox before being diluted to 2 × 106 cells/ml (t0) in synthetic medium without Dox. (Lower) GA-1020 cells carrying the empty vector (V), or pCM190-Dbf4-248, were grown in media with Dox, before a 2-h induction of cell cycle arrest by α-factor or nocodazole on media -Dox (2). Cells were then released into media without Dox and without inhibitors, and growth rates were determined by using a Casy cell counter. (C) FACS analysis of GA-850 cells transformed either with vector alone, or expressing either Dbf4-109 or Dbf4-248 fragments at t0 (G0 cells) or after 4 or 8 h outgrowth in the absence of drug, from a stationary culture. (D) pCM-190 (V) and Dbf4 fragment expression vectors described in Fig. 1 were used to transform _rad53_Δ _sml1_Δ (GA-1701), _mec1_Δ _sml1_Δ (GA-1702), _tel1_Δ (GA-856), and _chk1_Δ (GA-2012) strains. These were streaked out along with GA-850 transformants (WT), on plates without uracil and Dox , as in A. (E) Rad53p phosphorylation (P*) response in cells expressing Dbf4-248 fragment (−Dox) with or without 0.2 M HU is monitored by SDS/PAGE. GA-1040 (Rad53-13Myc tagged) was transformed with pCM190-Dbf4-248. Cells were cultured in SD medium without uracil and with 3 μg/ml Dox, and released into media lacking the drug for indicated times. Where indicated, 0.2 M HU was added.
Fig 4.
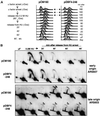
Dbf4p N-terminal fragment overexpression results in deregulation of late-origin firing after HU arrest. (A) GA-1040 cells transformed with either pCM190 or pCM-Dbf4-248 were cultured in SD medium without uracil and with 3 μg/ml Dox and were arrested with α-factor for 1.2 h. During α-factor arrest, Dbf4 fragments were induced for 45 min by removing Dox. Cells were harvested and resuspended in SD medium without uracil and without Dox, but containing 0.2 M HU. After 1 h**,** cells were centrifuged and resuspended in SD medium without uracil and without Dox, and progression through S phase was monitored by FACS analysis at the indicated times. An asynchronous culture of the same cells (Asy) and the samples arrested in pheromone (αF) or HU were also analyzed. (B) The samples taken up to 80 min after release into HU were examined by 2D gel analysis as described in Methods. Probes specific for the early ARS607 and late ARS603 were created by PCR. Open arrowheads indicate weak bubble arcs resulting from the initiation of replication, whereas the filled arrowhead indicates efficient initiation events.
Fig 5.
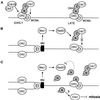
Model for the role of the Dbf4-Rad53p and Dbf4-ORC interactions in the control of late-firing origins. (A) Under normal conditions, Dbf4p targets Cdc7 to origins of replication through an interaction with ORC, and the recruitment or activation of Cdc28/Clb5 promotes origin firing. (B) Exposure to HU results in replication fork stalling and activation of Rad53p. Rad53p phosphorylates Dbf4p, preventing its association with late replication origins, and attenuating kinase activity (3). During recovery, Dbf4 is dephosphorylated. We propose that it then rebinds ORC and phosphorylates Mcm2p, leading to initiation at late origins. We do not know whether the Dbf4N-ORC interaction mutually excludes binding to Rad53p. (C) If HU arrest occurs in the presence of overexpressed Dbf4-N terminus, its binding to Rad53p and ORC may interfere with the proper activation of the checkpoint and recovery after HU removal. Consistent with the idea that phosphorylated Dbf4p may signal to downstream checkpoint targets, we suggest here that it activates Chk1p kinase to impair mitotic progression.
Similar articles
- A Dbf4p BRCA1 C-terminal-like domain required for the response to replication fork arrest in budding yeast.
Gabrielse C, Miller CT, McConnell KH, DeWard A, Fox CA, Weinreich M. Gabrielse C, et al. Genetics. 2006 Jun;173(2):541-55. doi: 10.1534/genetics.106.057521. Epub 2006 Mar 17. Genetics. 2006. PMID: 16547092 Free PMC article. - Association of RPA with chromosomal replication origins requires an Mcm protein, and is regulated by Rad53, and cyclin- and Dbf4-dependent kinases.
Tanaka T, Nasmyth K. Tanaka T, et al. EMBO J. 1998 Sep 1;17(17):5182-91. doi: 10.1093/emboj/17.17.5182. EMBO J. 1998. PMID: 9724654 Free PMC article. - ORC and the intra-S-phase checkpoint: a threshold regulates Rad53p activation in S phase.
Shimada K, Pasero P, Gasser SM. Shimada K, et al. Genes Dev. 2002 Dec 15;16(24):3236-52. doi: 10.1101/gad.239802. Genes Dev. 2002. PMID: 12502744 Free PMC article. - Stepwise assembly of initiation complexes at budding yeast replication origins during the cell cycle.
Diffley JF, Cocker JH, Dowell SJ, Harwood J, Rowley A. Diffley JF, et al. J Cell Sci Suppl. 1995;19:67-72. doi: 10.1242/jcs.1995.supplement_19.9. J Cell Sci Suppl. 1995. PMID: 8655649 Review. - Mechanisms involved in regulating DNA replication origins during the cell cycle and in response to DNA damage.
Early A, Drury LS, Diffley JF. Early A, et al. Philos Trans R Soc Lond B Biol Sci. 2004 Jan 29;359(1441):31-8. doi: 10.1098/rstb.2003.1362. Philos Trans R Soc Lond B Biol Sci. 2004. PMID: 15065654 Free PMC article. Review.
Cited by
- CDC5 inhibits the hyperphosphorylation of the checkpoint kinase Rad53, leading to checkpoint adaptation.
Vidanes GM, Sweeney FD, Galicia S, Cheung S, Doyle JP, Durocher D, Toczyski DP. Vidanes GM, et al. PLoS Biol. 2010 Jan 26;8(1):e1000286. doi: 10.1371/journal.pbio.1000286. PLoS Biol. 2010. PMID: 20126259 Free PMC article. - ATR/Mec1 prevents lethal meiotic recombination initiation on partially replicated chromosomes in budding yeast.
Blitzblau HG, Hochwagen A. Blitzblau HG, et al. Elife. 2013 Oct 1;2:e00844. doi: 10.7554/eLife.00844. Elife. 2013. PMID: 24137535 Free PMC article. - Incorporation into the prereplicative complex activates the Mcm2-7 helicase for Cdc7-Dbf4 phosphorylation.
Francis LI, Randell JC, Takara TJ, Uchima L, Bell SP. Francis LI, et al. Genes Dev. 2009 Mar 1;23(5):643-54. doi: 10.1101/gad.1759609. Genes Dev. 2009. PMID: 19270162 Free PMC article. - Mdt1, a novel Rad53 FHA1 domain-interacting protein, modulates DNA damage tolerance and G(2)/M cell cycle progression in Saccharomyces cerevisiae.
Pike BL, Yongkiettrakul S, Tsai MD, Heierhorst J. Pike BL, et al. Mol Cell Biol. 2004 Apr;24(7):2779-88. doi: 10.1128/MCB.24.7.2779-2788.2004. Mol Cell Biol. 2004. PMID: 15024067 Free PMC article. - An FHA domain-mediated protein interaction network of Rad53 reveals its role in polarized cell growth.
Smolka MB, Chen SH, Maddox PS, Enserink JM, Albuquerque CP, Wei XX, Desai A, Kolodner RD, Zhou H. Smolka MB, et al. J Cell Biol. 2006 Dec 4;175(5):743-53. doi: 10.1083/jcb.200605081. Epub 2006 Nov 27. J Cell Biol. 2006. PMID: 17130285 Free PMC article.
References
- Diffley J. F. X. (1998) Curr. Biol. 8, R771-R773. - PubMed
- Friedman K. L., Diller, J. D., Ferguson, B. M., Nyland, S. V., Brewer, B. J. & Fangman, W. L. (1996) Genes Dev. 10, 1595-1607. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases