Role for radA/sms in recombination intermediate processing in Escherichia coli - PubMed (original) (raw)
Role for radA/sms in recombination intermediate processing in Escherichia coli
Cynthia E Beam et al. J Bacteriol. 2002 Dec.
Abstract
RadA/Sms is a highly conserved eubacterial protein that shares sequence similarity with both RecA strand transferase and Lon protease. We examined mutations in the radA/sms gene of Escherichia coli for effects on conjugational recombination and sensitivity to DNA-damaging agents, including UV irradiation, methyl methanesulfonate (MMS), mitomycin C, phleomycin, hydrogen peroxide, and hydroxyurea (HU). Null mutants of radA were modestly sensitive to the DNA-methylating agent MMS and to the DNA strand breakage agent phleomycin, with conjugational recombination decreased two- to threefold. We combined a radA mutation with other mutations in recombination genes, including recA, recB, recG, recJ, recQ, ruvA, and ruvC. A radA mutation was strongly synergistic with the recG Holliday junction helicase mutation, producing profound sensitivity to all DNA-damaging agents tested. Lesser synergy was noted between a mutation in radA and recJ, recQ, ruvA, ruvC, and recA for sensitivity to various genotoxins. For survival after peroxide and HU exposure, a radA mutation surprisingly suppressed the sensitivity of recA and recB mutants, suggesting that RadA may convert some forms of damage into lethal intermediates in the absence of these functions. Loss of radA enhanced the conjugational recombination deficiency conferred by mutations in Holliday junction-processing function genes, recG, ruvA, and ruvC. A radA recG ruv triple mutant had severe recombinational defects, to the low level exhibited by recA mutants. These results establish a role for RadA/Sms in recombination and recombinational repair, most likely involving the stabilization or processing of branched DNA molecules or blocked replication forks because of its genetic redundancy with RecG and RuvABC.
Figures
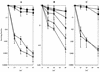
FIG. 1.
Synergy of radA with mutations affecting recombinational UV repair. Shown are UV survival curves for AB1157-derived strains both singly and doubly deficient for radA, recA, recJ, recQ, and recB. (A) AB1157, rec+ (▪); STL5280, radA1::kan (•); JC10287, recA_Δ (⧫); STL4799, recA_Δ radA1::kan (◊); (B) AB1157, rec+ (▪); STL5280, radA1::kan (•); STL1548, recQ1802::Tn_3 (▴); JC12123, recJ284::Tn_10 (⧫); STL5048, recQ1802::Tn_3 radA1_::kan (Δ); STL5042, recJ284::Tn_10 radA1_::kan (◊); (C) AB1157, rec+ (▪); STL5280, radA1::kan (•); N2101, recB268::Tn_10_ (▴); STL5480, recB268::Tn_10 radA1_::kan (Δ). Error bars indicate standard errors of the determinations.
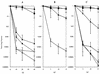
FIG. 2.
RadA is synergistic with Holliday junction-processing genes in UV repair. UV survival curves are shown for E. coli strains both singly and multiply deficient for radA, recG, ruvA, and ruvC. All strains assayed for UV survival were derived from the AB1157 background. (A) AB1157, rec+ (▪); STL5280, radA1::kan (•); N2096, ruvAΔ63 (▴); CS140, ruvC53 (⧫); STL5046, ruvC53 radA1::kan (◊); STL5037, ruvAΔ63 radA1::kan (Δ); (B) AB1157, rec+ (▪); STL5280, radA1::kan (•); N4452, recGΔ265::cat (▴); STL6588, recGΔ265::cat radA1::kan (Δ); (C) AB1157, rec+ (▪); STL5280, radA1::kan (•); STL6586, recGΔ265::cat ruvC53 (◊); STL6640, recGΔ265::cat radA1::kan ruvC53 (⧫); STL6571, recGΔ265::cat ruvAΔ63(Δ); STL6592, recGΔ265::cat radA1::kan ruvAΔ63 (▴). Error bars indicate standard errors of the determinations.
FIG. 3.
Double-strand-break (DSB)-mediated recombination. A broken fork can be repaired by recombinational reactions. (Likewise, ends of conjugative DNA or transducing fragments can be integrated via this mechanism.) Double-strand ends are resected by RecBCD nuclease; RecBCD also assists in loading of RecA onto single-strand DNA. The RecA-single-strand DNA filament promotes strand invasion into a homologous duplex molecule (the sister chromosome), forming a D-loop intermediate. Branch migration helicases can extend the region of pairing to form a Holliday junction (HJ), which can be resolved by cleavage mediated by Holliday junction endonucleases such as RuvC. Ligation of strand scissions restores an intact recombinant chromosome. RadA may participate in recombination by stabilizing any of these joint intermediates or by mediating branch migration or cleavage.
FIG. 4.
Fork regression and repair. Lesions can stall DNA polymerase. Fork regression catalyzed by RecG or RuvAB helicase activity can move the lesion into double-strand DNA, where it can be repaired. 5′ to 3′ degradation of the lagging strand by RecJ/RecQ may facilitate repair when the lagging strand has moved ahead of the leading strand. The regressed fork forms a Holliday junction, which can be cleaved by RuvC. Double-strand break repair can then restore integrity of the fork. Alternatively, RecBCD degradation of the double-strand arm or reversed branch migration can restore a fork structure.
FIG. 5.
Alignment of E. coli RadA/Sms with human Dmc1 showing similarity through the putative Zn finger region. Arrows indicate the conserved cysteines of the Zn finger.
Similar articles
- Genetic analysis of Escherichia coli RadA: functional motifs and genetic interactions.
Cooper DL, Boyle DC, Lovett ST. Cooper DL, et al. Mol Microbiol. 2015 Mar;95(5):769-79. doi: 10.1111/mmi.12899. Epub 2015 Jan 30. Mol Microbiol. 2015. PMID: 25484163 Free PMC article. - Replication arrest-stimulated recombination: Dependence on the RecA paralog, RadA/Sms and translesion polymerase, DinB.
Lovett ST. Lovett ST. DNA Repair (Amst). 2006 Dec 9;5(12):1421-7. doi: 10.1016/j.dnarep.2006.06.008. Epub 2006 Aug 9. DNA Repair (Amst). 2006. PMID: 16904387 - Roles of the recG gene product of Escherichia coli in recombination repair: effects of the delta recG mutation on cell division and chromosome partition.
Ishioka K, Iwasaki H, Shinagawa H. Ishioka K, et al. Genes Genet Syst. 1997 Apr;72(2):91-9. doi: 10.1266/ggs.72.91. Genes Genet Syst. 1997. PMID: 9265736 - Biological roles of the Escherichia coli RuvA, RuvB and RuvC proteins revealed.
West SC, Connolly B. West SC, et al. Mol Microbiol. 1992 Oct;6(19):2755-9. doi: 10.1111/j.1365-2958.1992.tb01454.x. Mol Microbiol. 1992. PMID: 1435254 Review. - Processing of Holliday junctions by the Escherichia coli RuvA, RuvB, RuvC and RecG proteins.
Müller B, West SC. Müller B, et al. Experientia. 1994 Mar 15;50(3):216-22. doi: 10.1007/BF01924004. Experientia. 1994. PMID: 8143795 Review.
Cited by
- Radiation-sensitive gene A (RadA) targets DisA, DNA integrity scanning protein A, to negatively affect cyclic Di-AMP synthesis activity in Mycobacterium smegmatis.
Zhang L, He ZG. Zhang L, et al. J Biol Chem. 2013 Aug 2;288(31):22426-36. doi: 10.1074/jbc.M113.464883. Epub 2013 Jun 10. J Biol Chem. 2013. PMID: 23760274 Free PMC article. - Identification of recG genetic interactions in Escherichia coli by transposon sequencing.
Bonde NJ, Wood EA, Myers KS, Place M, Keck JL, Cox MM. Bonde NJ, et al. J Bacteriol. 2023 Dec 19;205(12):e0018423. doi: 10.1128/jb.00184-23. Epub 2023 Nov 29. J Bacteriol. 2023. PMID: 38019006 Free PMC article. - RADA-dependent branch migration has a predominant role in plant mitochondria and its defect leads to mtDNA instability and cell cycle arrest.
Chevigny N, Weber-Lotfi F, Le Blevenec A, Nadiras C, Fertet A, Bichara M, Erhardt M, Dietrich A, Raynaud C, Gualberto JM. Chevigny N, et al. PLoS Genet. 2022 May 12;18(5):e1010202. doi: 10.1371/journal.pgen.1010202. eCollection 2022 May. PLoS Genet. 2022. PMID: 35550632 Free PMC article. - Metagenomic analysis of 0.2-μm-passable microorganisms in deep-sea hydrothermal fluid.
Nakai R, Abe T, Takeyama H, Naganuma T. Nakai R, et al. Mar Biotechnol (NY). 2011 Oct;13(5):900-8. doi: 10.1007/s10126-010-9351-6. Epub 2011 Jan 29. Mar Biotechnol (NY). 2011. PMID: 21279410 - Classical and novel properties of Holliday junction resolvase SynRuvC from Synechocystis sp. PCC6803.
Gu Y, Yang Y, Kou C, Peng Y, Yang W, Zhang J, Jin H, Han X, Wang Y, Shen X. Gu Y, et al. Front Microbiol. 2024 Apr 18;15:1362880. doi: 10.3389/fmicb.2024.1362880. eCollection 2024. Front Microbiol. 2024. PMID: 38699476 Free PMC article.
References
- Bachmann, B. J. 1996. Derivations and genotypes of some mutant derivatives of Escherichia coli K-12, p. 2460-2488. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd., vol. 2. ASM Press, Washington, D.C.
- Berg, J. M. 1986. Potential metal-binding domains in nucleic acid binding proteins. Science 232:485-487. - PubMed
- Bishop, D. K., D. Park, L. Xu, and N. Kleckner. 1992. DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell 69:439-456. - PubMed
- Byfield, J. E., Y. C. Lee, L. Tu, and F. Kulhanian. 1976. Molecular interactions of the combined effects of bleomycin and x-rays on mammalian cell survival. Cancer Res. 36:1138-1143. - PubMed
- Carrasco, B., S. Fernandez, K. Asai, N. Ogasawara, and C. Alonso. 2002. Effect of the recU suppressors sms and subA on DNA repair and homologous recombination in Bacillus subtilis. Mol. Genet. Genomics 266:899-906. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM051753/GM/NIGMS NIH HHS/United States
- T32 GM007122/GM/NIGMS NIH HHS/United States
- R01 GM51753/GM/NIGMS NIH HHS/United States
- T32 GM07122/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases