Sequencing-independent method to generate oligonucleotide probes targeting a variable region in bacterial 16S rRNA by PCR with detachable primers - PubMed (original) (raw)
Sequencing-independent method to generate oligonucleotide probes targeting a variable region in bacterial 16S rRNA by PCR with detachable primers
Stefan Bertilsson et al. Appl Environ Microbiol. 2002 Dec.
Abstract
Oligonucleotide probes targeting the small-subunit rRNA are commonly used to detect and quantify bacteria in natural environments. We developed a PCR-based approach that allows synthesis of oligonucleotide probes targeting a variable region in the 16S rRNA without prior knowledge of the target sequence. Analysis of all 16S rRNA gene sequences in the Ribosomal Database Project database revealed two universal primer regions bracketing a variable, population-specific region. The probe synthesis is based on a two-step PCR amplification of this variable region in the 16S rRNA gene by using three universal bacterial primers. First, a double-stranded product is generated, which then serves as template in a linear amplification. After each of these steps, products are bound to magnetic beads and the primers are detached through hydrolysis of a ribonucleotide at the 3' end of the primers. This ultimately produces a single-stranded oligonucleotide of about 30 bases corresponding to the target. As probes, the oligonucleotides are highly specific and could discriminate between nucleic acids from closely and distantly related bacterial strains, including different species of VIBRIO: The method will facilitate rapid generation of oligonucleotide probes for large-scale hybridization assays such as screening of clone libraries or strain collections, ribotyping microarrays, and in situ hybridization. An additional advantage of the method is that fluorescently or radioactively labeled nucleotides can be incorporated during the second amplification, yielding intensely labeled probes.
Figures
FIG. 1.
Neighbor-joining tree of 16S rRNAs for a collection of 44 bacterial isolates from a coastal marine environment, reference strains, and sequences retrieved from GenBank. The scale bar indicates 10% sequence difference. Organism names for the isolates are from the closest matches in GenBank. Group affiliations, GenBank accession numbers, and isolate-specific target sequences for PCR-generated 16S rRNA oligonucleotide probes are shown for each sequence.
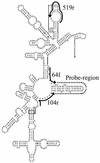
FIG. 2.
Partial secondary structure of 16S rRNA from E. coli (4), with target regions for the three Bacteria universal primers (64f, 104r, and 519r) used in the probe synthesis indicated by solid lines. The dashed line indicates the variable probing region.
FIG. 3.
Outline of PCR-based method to generate population-specific oligonucleotide probes without prior sequence information.
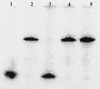
FIG. 4.
(A) Phosphorimaging scan showing detachment of the 5′-32P-labeled primer from the PCR product after alkaline hydrolysis of the ribonucleotide at the 3′ end of primer 104r. PCR products with primer 104r lacking the 3′ ribonucleotides are included for comparison. Lanes: 1, primer 104r; 2, PCR product with 3′ ribonucleotide-modified primer; 3, PCR product with 3′ ribonucleotide-modified primer after alkaline hydrolysis; 4, PCR product with nonmodified primer; 5, PCR product with nonmodified primer after alkaline hydrolysis.
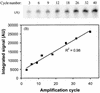
FIG. 5.
Linear amplification with 5′-32P-labeled primer 104r. (A) Gel electrophoresis and phosphorimaging detection of amplified fragments after 3 to 40 cycles. (B) Quantification by image analysis using local background subtraction shows a linear accumulation of amplification product with cycle number. AU, absorbance units.
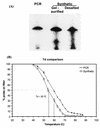
FIG. 6.
(A) Gel electrophoresis of a 5′-32P-labeled PCR-produced oligonucleotide probe targeting 16S rRNA of E. coli. Two synthetic oligonucleotides of identical sequence (desalted or purified by acrylamide gel electrophoresis) are included as a reference. (B) Dissociation curves for a commercially produced, gel-purified oligonucleotide probe and a PCR-generated oligonucleotide probe with identical sequence (5′ACTCGTCAGCAAAGAGCAAGCTTCTTCCTGTTACCGTT3′). Each symbol represents the mean dissociation of probe hybridized to triplicate slot blots with total nucleic acids from E. coli as a target (∼50 ng of 16S rRNA). Error bars represent ±1 standard deviation, and dashed lines indicate the Td for the two probes.
FIG. 7.
Membrane hybridization of total nucleic acids from 30 bacterial strains with 32P-labeled oligonucleotide probes. (A) Identity and location of immobilized nucleic acids on the membrane; (B) hybridizations with universal bacterial probe EUB338; (C) PCR-generated probe specific for E. coli; (D) PCR-generated probe specific for V. anguillarum.
Similar articles
- PCR primers and probes for the 16S rRNA gene of most species of pathogenic bacteria, including bacteria found in cerebrospinal fluid.
Greisen K, Loeffelholz M, Purohit A, Leong D. Greisen K, et al. J Clin Microbiol. 1994 Feb;32(2):335-51. doi: 10.1128/jcm.32.2.335-351.1994. J Clin Microbiol. 1994. PMID: 7512093 Free PMC article. - Use of subtractive hybridization to design habitat-based oligonucleotide probes for investigation of natural bacterial communities.
Mau M, Timmis KN. Mau M, et al. Appl Environ Microbiol. 1998 Jan;64(1):185-91. doi: 10.1128/AEM.64.1.185-191.1998. Appl Environ Microbiol. 1998. PMID: 9435075 Free PMC article. - Development of PCR-based hybridization protocol for identification of streptococcal species.
Bentley RW, Leigh JA. Bentley RW, et al. J Clin Microbiol. 1995 May;33(5):1296-301. doi: 10.1128/jcm.33.5.1296-1301.1995. J Clin Microbiol. 1995. PMID: 7542267 Free PMC article. - Review and re-analysis of domain-specific 16S primers.
Baker GC, Smith JJ, Cowan DA. Baker GC, et al. J Microbiol Methods. 2003 Dec;55(3):541-55. doi: 10.1016/j.mimet.2003.08.009. J Microbiol Methods. 2003. PMID: 14607398 Review. - Phylogenetic identification and in situ detection of individual microbial cells without cultivation.
Amann RI, Ludwig W, Schleifer KH. Amann RI, et al. Microbiol Rev. 1995 Mar;59(1):143-69. doi: 10.1128/mr.59.1.143-169.1995. Microbiol Rev. 1995. PMID: 7535888 Free PMC article. Review.
Cited by
- System Integration - A Major Step toward Lab on a Chip.
Sin ML, Gao J, Liao JC, Wong PK. Sin ML, et al. J Biol Eng. 2011 May 25;5:6. doi: 10.1186/1754-1611-5-6. J Biol Eng. 2011. PMID: 21612614 Free PMC article. - Response of Tomato Rhizosphere Bacteria to Root-Knot Nematodes, Fenamiphos and Sampling Time Shows Differential Effects on Low Level Taxa.
Colagiero M, Rosso LC, Catalano D, Schena L, Ciancio A. Colagiero M, et al. Front Microbiol. 2020 Mar 20;11:390. doi: 10.3389/fmicb.2020.00390. eCollection 2020. Front Microbiol. 2020. PMID: 32265860 Free PMC article. - A detailed analysis of 16S ribosomal RNA gene segments for the diagnosis of pathogenic bacteria.
Chakravorty S, Helb D, Burday M, Connell N, Alland D. Chakravorty S, et al. J Microbiol Methods. 2007 May;69(2):330-9. doi: 10.1016/j.mimet.2007.02.005. Epub 2007 Feb 22. J Microbiol Methods. 2007. PMID: 17391789 Free PMC article. - Legacy Metal Contaminants and Excess Nutrients in Low Flow Estuarine Embayments Alter Composition and Function of Benthic Bacterial Communities.
Birrer SC, Wemheuer F, Dafforn KA, Gribben PE, Steinberg PD, Simpson SL, Potts J, Scanes P, Doblin MA, Johnston EL. Birrer SC, et al. Front Microbiol. 2021 Oct 8;12:661177. doi: 10.3389/fmicb.2021.661177. eCollection 2021. Front Microbiol. 2021. PMID: 34690940 Free PMC article. - Differences in Intertidal Microbial Assemblages on Urban Structures and Natural Rocky Reef.
Tan EL, Mayer-Pinto M, Johnston EL, Dafforn KA. Tan EL, et al. Front Microbiol. 2015 Nov 20;6:1276. doi: 10.3389/fmicb.2015.01276. eCollection 2015. Front Microbiol. 2015. PMID: 26635747 Free PMC article.
References
- Gutell, R. R., J. J. Cannone, Z. Shang, Y. Du, and M. J. Serra. 2000. A story: unpaired adenosine bases in ribosomal RNA. J. Mol. Biol. 304:335-354. - PubMed
- Head, I. M., J. R. Saunders, and R. W. Pickup. 1998. Microbial evolution, diversity and ecology; a decade of ribosomal RNA analysis of uncultivated microorganisms. Microb. Ecol. 35:1-21. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials