Allele-specific histone lysine methylation marks regulatory regions at imprinted mouse genes - PubMed (original) (raw)
Allele-specific histone lysine methylation marks regulatory regions at imprinted mouse genes
Cécile Fournier et al. EMBO J. 2002.
Abstract
In different eukaryotic model systems, chromatin and gene expression are modulated by post-translational modification of histone tails. In this in vivo study, histone methylation and acetylation are investigated along the imprinted mouse genes Snrpn, Igf2r and U2af1-rs1. These imprinted genes all have a CpG-rich regulatory element at which methylation is present on the maternal allele, and originates from the female germ line. At these 'differentially methylated regions' (DMRs), histone H3 on the paternal allele has lysine-4 methylation and is acetylated. On the maternally inherited allele, in contrast, chromatin is marked by hypermethylation on lysine-9 of H3. Allele-specific patterns of lysine-4 and lysine-9 methylation are also detected at other regions of the imprinted loci. For the DMR at the U2af1-rs1 gene, we establish that the methyl-CpG-binding-domain (MBD) proteins MeCP2, MBD1 and MBD3 are associated with the maternal allele. These data support the hypothesis that MBD protein-associated histone deacetylase/chromatin-remodelling complexes are recruited to the parental allele that has methylated DNA and H3-K9 methylation, and are prevented from binding to the opposite allele by H3 lysine-4 methylation.
Figures
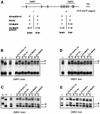
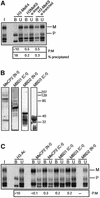
Fig. 1. Differential histone methylation and acetylation at Snrpn. (A) Schematic presentation of Snrpn showing exons (vertical bars) and DMRs (horizontal lines). Regions analysed by ChIP are indicated underneath (small horizontal bars). A summary of the maternal (M) and/or paternal (P) allele specificity of the precipitated chromatin is presented underneath for ChIP assays on H3 acetylation on lysines 9 and 14 (H3-AcK9+14), H4 acetylated at lysines 5, 8, 12 and 16 (H4-Ac), H3 methylated at K4 (H3-MeK4) and H3 methylated at K9 (H3-MeK9). (B) ChIP at the DMR1 in brain. Chromatin was extracted from brain tissue dissected from mice that were (C57BL/6 × M.spretus) F1 for chromosome 7, and was precipitated with antisera to H3-AcK9+14, H4-Ac, H3-MeK4 and H3-MeK9. DNAs extracted from antibody-bound (B) and unbound (U) fractions were used for amplification at the DMR1. PCR products were denatured and run through an SSCP gel. Maternal (M) and paternal (P) bands are indicated. To the left, PCR products from control C57BL/6 (D) and M.spretus (S) genomic DNAs and from the input chromatin (I) used for ChIP. (C) The DMR2 in brain. The same bound and unbound fractions as in panel B were used for PCR amplification from the DMR2. (D) The DMR1 in liver. Liver chromatin was precipitated with different antisera, and extracted DNA samples were amplified with the primer pair from the DMR1. (E) The DMR2 in liver. The same extracted DNA samples as in panel D were used as template for amplification with primers from the DMR2.
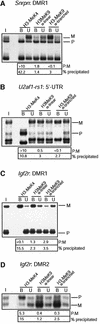
Fig. 2. H3-K9 hypermethylation on chromatin associated with the DNA-methylated allele of DMRs. Liver chromatin was precipitated with antisera to H3-K4 dimethylation (H3-MeK4), to a linear peptide with K9 dimethylation (α-linear) and to a branched peptide with K9 dimethylation (α-branched). (A) Maternal K9 methylation at the Snrpn imprinting-control centre. DNAs extracted from bound and unbound fractions were amplified with primers from the Snrpn DMR1; to the left, amplification from the input chromatin (I). P:M ratios (corrected for P:M in the input chromatin) are indicated. Real-time PCR was applied on the bound fractions to determine the percentage of chromatin precipitated. (B) Maternal K9 methylation at U2af1-rs1. PCR was with the primers from the 5′-UTR. (C) Paternal K9 methylation at the Igf2r DMR1 region. PCR was with the primers from the DMR1. (D) Maternal K9 methylation at the DMR2 of Igf2r. Amplification was with the DMR2 primers.
Fig. 3. Allele-specific histone methylation at the Igf2r gene. (A) Schematic presentation of the Igf2r gene, showing the DMR1 (paternal DNA methyl ation), DMR2 (maternal DNA methylation), the transcription initiation sites for sense and antisense transcription (arrows), and the position of exons (filled boxes). Regions analysed by ChIP are indicated underneath (small horizontal bars). (B) The DMR1 in liver. Presentation of results is as in Figure 1. (C) The DMR2 in liver. The same samples were analysed by PCR amplification with the primers from the DMR2. (D) 3′-end of the gene. The same samples as in Figure 2B were analysed by PCR amplification with primers at exon 48. (E) The DMR1 in brain. Chromatin extracted from brain was immunoprecipitated and analysed with the primers from the DMR1. (F) The DMR2 in brain. The precipitated samples as in Figure 2C were analysed with the primers at the DMR2. (G) 3′-end of the gene. The same samples as in Figure 2B were analysed by PCR amplification with primers at exon 48.
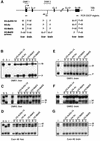
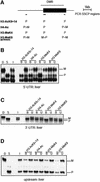
Fig. 4. Paternal H3 acetylation and K4 methylation at U2af1-rs1. (A) Schematic presentation of the intronless U2af1-rs1 gene (filled box) with the DMR indicated above. Regions analysed by ChIP are indicated underneath (small horizontal bars). (B) The 5′-UTR. Chromatin extracted from liver was precipitated with different antisera, and after extraction of DNA from the bound and unbound fractions, amplification was with primers from the 5′-UTR. (C) The 3′-UTR. The same precipitation samples and control DNAs as in (B) were used for amplification with a primer pair from the 3′-UTR. (D) Chromatin upstream of U2af1-rs1. The same DNA samples as in (B) were used for amplification with a primer pair from a region at 2 kb upstream of the gene.
Fig. 5. MBD proteins associate with the maternal allele of U2af1-rs1. (A) Analysis of H3-K9 methylation on crosslinked chromatin. Formaldehyde crosslinked chromatin was precipitated with the α-linear and α-branched antisera to H3-MeK9. DNA from the bound and unbound fractions was amplified with primers from the 5′-UTR. To the left, amplification from input chromatin (I). P:M ratios, corrected for P:M in the input chromatin, are indicated, as well as the percentage of chromatin precipitated at U2af1-rs1. (B) The newly derived antisera αMeCP2 N-t, αMBD1 C-t, αMBD2 N-t and αMBD3 C-t were tested on a western blot with total nuclear extract from a murine cell line. The density markers indicate a pre-stained standard (Kaleidoscope, Bio-Rad). Comparable results were obtained with a human cell line (data not shown). (C) Crosslinked chromatin was precipitated with antisera to MeCP2, MBD1 and MBD3. Corrected P:M ratios are indicated for the precipitated fractions.
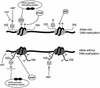
Fig. 6. A working model for the maintenance of opposite epigenetic states at DMRs. Methylation (M) of DNA is present on one of the parental alleles only (at many DMRs, this is the maternal allele). This methylation is recognized by MBD proteins, which recruit HDAC/chromatin-remodelling complexes to the chromatin. HDACs (filled circles) ensure deacetylation of histones and may be associated with DNMTs, which maintain the DNA methylated. MBD protein binding may also be mechanistically linked to the H3-K9 hypermethylation on this parental allele, and this could constitute a second element in the maintenance of its DNA methylation. The opposite parental allele has no DNA methylation and is not accessible to HDAC complexes and associated DNMT activities, possibly in part because of the H3-K4 methylation. H3-K4 methylation could thus prevent DNA methylation from becoming established. In contrast, HATs and K4 histone methyltransferase (HMT) have access to the chromatin and this maintains the acetylation and K4 methylation on this allele.
Similar articles
- DNA methylation is linked to deacetylation of histone H3, but not H4, on the imprinted genes Snrpn and U2af1-rs1.
Gregory RI, Randall TE, Johnson CA, Khosla S, Hatada I, O'Neill LP, Turner BM, Feil R. Gregory RI, et al. Mol Cell Biol. 2001 Aug;21(16):5426-36. doi: 10.1128/MCB.21.16.5426-5436.2001. Mol Cell Biol. 2001. PMID: 11463825 Free PMC article. - Inhibition of histone deacetylases alters allelic chromatin conformation at the imprinted U2af1-rs1 locus in mouse embryonic stem cells.
Gregory RI, O'Neill LP, Randall TE, Fournier C, Khosla S, Turner BM, Feil R. Gregory RI, et al. J Biol Chem. 2002 Apr 5;277(14):11728-34. doi: 10.1074/jbc.M105775200. Epub 2002 Jan 30. J Biol Chem. 2002. PMID: 11821379 - Tissue-specific and imprinted epigenetic modifications of the human NDN gene.
Lau JC, Hanel ML, Wevrick R. Lau JC, et al. Nucleic Acids Res. 2004 Jun 24;32(11):3376-82. doi: 10.1093/nar/gkh671. Print 2004. Nucleic Acids Res. 2004. PMID: 15247330 Free PMC article. - Cross-species clues of an epigenetic imprinting regulatory code for the IGF2R gene.
Vu TH, Jirtle RL, Hoffman AR. Vu TH, et al. Cytogenet Genome Res. 2006;113(1-4):202-8. doi: 10.1159/000090833. Cytogenet Genome Res. 2006. PMID: 16575181 Review. - [The roles of histone lysine methylation in epigenetic regulation].
Du TT, Huang QH. Du TT, et al. Yi Chuan. 2007 Apr;29(4):387-92. doi: 10.1360/yc-007-0387. Yi Chuan. 2007. PMID: 17548299 Review. Chinese.
Cited by
- Identification of the imprinted KLF14 transcription factor undergoing human-specific accelerated evolution.
Parker-Katiraee L, Carson AR, Yamada T, Arnaud P, Feil R, Abu-Amero SN, Moore GE, Kaneda M, Perry GH, Stone AC, Lee C, Meguro-Horike M, Sasaki H, Kobayashi K, Nakabayashi K, Scherer SW. Parker-Katiraee L, et al. PLoS Genet. 2007 May 4;3(5):e65. doi: 10.1371/journal.pgen.0030065. Epub 2007 Mar 12. PLoS Genet. 2007. PMID: 17480121 Free PMC article. - Differential histone modifications mark mouse imprinting control regions during spermatogenesis.
Delaval K, Govin J, Cerqueira F, Rousseaux S, Khochbin S, Feil R. Delaval K, et al. EMBO J. 2007 Feb 7;26(3):720-9. doi: 10.1038/sj.emboj.7601513. Epub 2007 Jan 25. EMBO J. 2007. PMID: 17255950 Free PMC article. - Origin and Evolution of Marsupial-specific Imprinting Clusters Through Lineage-specific Gene Duplications and Acquisition of Promoter Differential Methylation.
Cao W, Douglas KC, Samollow PB, VandeBerg JL, Wang X, Clark AG. Cao W, et al. Mol Biol Evol. 2023 Feb 3;40(2):msad022. doi: 10.1093/molbev/msad022. Mol Biol Evol. 2023. PMID: 36721950 Free PMC article. - Allele-specific transcriptional elongation regulates monoallelic expression of the IGF2BP1 gene.
Thomas BJ, Rubio ED, Krumm N, Broin PO, Bomsztyk K, Welcsh P, Greally JM, Golden AA, Krumm A. Thomas BJ, et al. Epigenetics Chromatin. 2011 Aug 3;4:14. doi: 10.1186/1756-8935-4-14. Epigenetics Chromatin. 2011. PMID: 21812971 Free PMC article. - Establishing, maintaining and modifying DNA methylation patterns in plants and animals.
Law JA, Jacobsen SE. Law JA, et al. Nat Rev Genet. 2010 Mar;11(3):204-20. doi: 10.1038/nrg2719. Nat Rev Genet. 2010. PMID: 20142834 Free PMC article. Review.
References
- Arney K.L, Bao,S., Bannister,A.J., Kouzarides,T. and Surani,M.A. (2002) Histone methylation defines epigenetic asymmetry in the mouse zygote. Int. J. Dev. Biol., 46, 317–320. - PubMed
- Ballestar E. and Wolffe,A.P. (2001) Methyl-CpG-binding proteins. Targeting specific gene repression. Eur. J. Biochem., 268, 1–6. - PubMed
- Bannister A.J., Schneider,R. and Kouzarides,T. (2002) Histone methylation. Dynamic or static? Cell, 109, 801–806. - PubMed
- Bielinska B, Blaydes,S.M., Buiting,K., Yang,T., Krajewska-Walasek,M., Horsthemke,B. and Brannan,C.I. (2000) De novo deletions of SNRPN exon 1 in early human and mouse embryos result in a paternal to maternal imprint switch. Nat. Genet., 25, 74–78. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous