Structures of HIV-1 reverse transcriptase with pre- and post-translocation AZTMP-terminated DNA - PubMed (original) (raw)
. 2002 Dec 2;21(23):6614-24.
doi: 10.1093/emboj/cdf637.
Arthur D Clark Jr, Kalyan Das, Steve Tuske, Jens J Birktoft, Palanichamy Ilankumaran, Andagar R Ramesha, Jane M Sayer, Donald M Jerina, Paul L Boyer, Stephen H Hughes, Eddy Arnold
Affiliations
- PMID: 12456667
- PMCID: PMC136941
- DOI: 10.1093/emboj/cdf637
Structures of HIV-1 reverse transcriptase with pre- and post-translocation AZTMP-terminated DNA
Stefan G Sarafianos et al. EMBO J. 2002.
Abstract
AZT (3'-azido-3'-deoxythymidine) resistance involves the enhanced excision of AZTMP from the end of the primer strand by HIV-1 reverse transcriptase. This reaction can occur when an AZTMP-terminated primer is bound at the nucleotide-binding site (pre-translocation complex N) but not at the 'priming' site (post-translocation complex P). We determined the crystal structures of N and P complexes at 3.0 and 3.1 A resolution. These structures provide insight into the structural basis of AZTMP excision and the mechanism of translocation. Docking of a dNTP in the P complex structure suggests steric crowding in forming a stable ternary complex that should increase the relative amount of the N complex, which is the substrate for excision. Structural differences between complexes N and P suggest that the conserved YMDD loop is involved in translocation, acting as a springboard that helps to propel the primer terminus from the N to the P site after dNMP incorporation.
Figures
Fig. 1. Schematic representation of the mechanism of DNA polymerization indicating the proposed occupancy of sites P (‘
p
riming’ site, in blue) and N (‘
n
ucleotide-binding site’, in red) by the 3′ end of the primer during the course of the reaction. The structures reported here, RT(P)–AZTMP (complex P) and RT(N)–AZTMP (complex N), have AZTMP-terminated primers at the P and N sites, respectively. Step 1: binding of DNA to free enzyme E [RT(apo)] with the 3′ primer end at the P site. Step 2: binding of dNTP to the N site to form an ‘open’ ternary complex [open RT(ter)]. Step 3: formation of a ‘closed’ ternary complex [closed RT(ter)] through conformational change. Step 4: bond formation accompanied by release of pyrophosphate to form the non-translocated complex N [RT(N)–dNMP]. Step 5: in processive synthesis, the primer translocates from the N site to the P site. Step 5′: in non-processive synthesis, DNA dissociates from the enzyme.
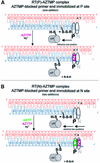
Fig. 2. Reaction schemes for cross-linking AZTMP-terminated template–primers at the P (priming) (A) or N (nucleotide-binding) (B) sites of HIV-1 RT. Cross-linking takes place when Cys258 of the αH helix of the p66 thumb of HIV-1 RT is aligned with the cross-linkable nucleotide analog. Complex P [RT(P)–AZTMP complex] is formed after addition of AZTMP (A), whereas complex N [RT(N)– AZTMP complex] is formed with in situ incorporation of dAMP and AZTMP (B). Z is AZTMP.
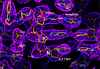
Fig. 3. Simulated annealing (2_F_obs – _F_calc, 1.2σ contours) omit electron density map at the polymerase active site region in complex N (omitting AZTMP and residues within a 5.5 Å radius).
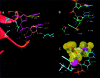
Fig. 4. Ribbon representation of superposed polymerase active sites of complexes P and N (alignment based on p66 residues 107–112 and 155–215). Color scheme: primer strands of complex P (green) and N (magenta), palm subdomains of complexes P (gray) and N (red), and side chains of complexes P (gray) and N (magenta).
Fig. 5. (A) Ribbon representation of superposed polymerase active sites (same basis of superposition as used for Figure 4) of complex N and HIV-1 RT/DNA/dNTP ternary complex [RT(ter)–ddNMP–dTTP complex] (Huang et al., 1998); PDB code 1RTD. Color scheme: side chains of complex N (cyan), primer strand of complex N (magenta), incoming dNTP of the ternary complex (yellow), metals A and B in the ternary complex (yellow). In complex N, the corresponding metals (A′ and B′) are either not seen in the structure and may have been released together with PPi (A′), or are observed (Figure 3) at a position shifted by ∼4.7 Å (metal B′). (B) Superposition of polymerase active sites of the non-terminated [RT(P)–dNMP, green] (Ding et al., 1998; PDB code 2HMI) and AZTMP-terminated P complex [RT(P)–AZTMP, white]. The main structural difference is in the inclination of the terminal nucleotide. (C) Superposition of the polymerase active sites (aligned using p66 residues 107–112 and 155–215) of complex P (in white) on the RT(ter)–ddNMP/dTTP ternary complex (in cyan) (Huang et al., 1998); PDB code 1RTD. The ternary complex YMDD loop is displaced ∼1.0 Å from its position in the P complex. Steric conflicts (in red) are mostly between the C5′ of the incoming dNTP and the side chain of Asp185.
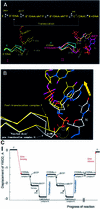
Fig. 6. (A) Conformations of conserved polymerase active site YMDD motif of RT during polymerization. Cα traces of the unliganded [RT(apo), cyan] (Hsiou et al., 1996), complex P [RT(P)–AZTMP, yellow], ternary [RT(ter)–ddNMP/dTTP, white] (Huang et al., 1998) and complex N [RT(N)–AZTMP, magenta] structures. In I, superposition (based on p66 residues 107–112 and 155–215) of unliganded RT(apo) and complex P reveals movement of the YMDD loop in the RT(P)–AZTMP complex relative to RT(apo). In II, superposition of the unliganded RT(apo), complex P [RT(P)–AZTMP] and ternary complex [RT(ter)–ddNMP/dTTP] polymerase active sites shows further displacement of the YMDD loop in the ternary complex. (B) Superposition of RT P and N complexes with AZTMP-terminated nucleic acids shows movement of the YMDD loop following translocation. The Cα of D185 is displaced ∼1.7 Å in complex P, ∼2.2 Å in the ternary complex and ∼2.7 Å in complex N from its reference position in unliganded RT (apo, cyan). (C) Displacement of the Cα carbon of Asp185 of the conserved YMDD motif relative to the unliganded structure of RT(apo) (PDB code: 1DLO). Negatives values in Å correspond to compression. At the lowest point [RT(N)–AZTMP complex], the displacement is ∼2.7 Å. Non-nucleoside reverse transcriptase inhibitor (NNRTI)-bound RT structures have displacements in the opposite direction (YMDD positive), suggesting that conformational changes of YMDD could influence inhibition by NNRTIs (Ding et al., 1998).
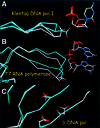
Fig. 7. Structural changes caused by active substrate binding in other processive (Klenow fragment of E.coli pol I and T7 RNA polymerase) and non-processive (human β-pol) polymerases. (A) Structures of the Klenow fragment of Thermus aquaticus DNA polymerase I in the presence (white) and absence (cyan) of nucleic acid substrate (PDB codes 1QTM and 1KTQ). (B) T7 RNA polymerase with nucleic acid present (PDB code 1CEZ) and absent (in cyan; PDB code 1ARO). The 1ARO structure has lysozyme inhibitor bound at a site other than the polymerase site. (C) Structures of human polymerase β in the presence (white) and absence (cyan) of nucleic acid substrate (PDB codes 1BPX and 1BPD).
Fig. 8. Schematic relationships among events that affect excision-based NRTI resistance (dNTP binding, translocation, excision). Factors that affect any stage will affect the overall equilibrium. X is an NRTI (red), A is ATP (orange) and dNTP (cyan) is the cognate nucleotide triphosphate.
Similar articles
- The M184V mutation reduces the selective excision of zidovudine 5'-monophosphate (AZTMP) by the reverse transcriptase of human immunodeficiency virus type 1.
Boyer PL, Sarafianos SG, Arnold E, Hughes SH. Boyer PL, et al. J Virol. 2002 Apr;76(7):3248-56. doi: 10.1128/jvi.76.7.3248-3256.2002. J Virol. 2002. PMID: 11884549 Free PMC article. - Nucleoside analog resistance caused by insertions in the fingers of human immunodeficiency virus type 1 reverse transcriptase involves ATP-mediated excision.
Boyer PL, Sarafianos SG, Arnold E, Hughes SH. Boyer PL, et al. J Virol. 2002 Sep;76(18):9143-51. doi: 10.1128/jvi.76.18.9143-9151.2002. J Virol. 2002. PMID: 12186898 Free PMC article. - Selective excision of AZTMP by drug-resistant human immunodeficiency virus reverse transcriptase.
Boyer PL, Sarafianos SG, Arnold E, Hughes SH. Boyer PL, et al. J Virol. 2001 May;75(10):4832-42. doi: 10.1128/JVI.75.10.4832-4842.2001. J Virol. 2001. PMID: 11312355 Free PMC article. - Structural requirements for efficient phosphorylation of nucleotide analogs by human thymidylate kinase.
Lavie A, Konrad M. Lavie A, et al. Mini Rev Med Chem. 2004 May;4(4):351-9. doi: 10.2174/1389557043403981. Mini Rev Med Chem. 2004. PMID: 15134538 Review.
Cited by
- Analysis of human immunodeficiency virus type 1 reverse transcriptase subunit structure/function in the context of infectious virions and human target cells.
Mulky A, Kappes JC. Mulky A, et al. Antimicrob Agents Chemother. 2005 Sep;49(9):3762-9. doi: 10.1128/AAC.49.9.3762-3769.2005. Antimicrob Agents Chemother. 2005. PMID: 16127051 Free PMC article. - Effects of the Delta67 complex of mutations in human immunodeficiency virus type 1 reverse transcriptase on nucleoside analog excision.
Boyer PL, Imamichi T, Sarafianos SG, Arnold E, Hughes SH. Boyer PL, et al. J Virol. 2004 Sep;78(18):9987-97. doi: 10.1128/JVI.78.18.9987-9997.2004. J Virol. 2004. PMID: 15331732 Free PMC article. - Impact of primer-induced conformational dynamics of HIV-1 reverse transcriptase on polymerase translocation and inhibition.
Auger A, Beilhartz GL, Zhu S, Cauchon E, Falgueyret JP, Grobler JA, Ehteshami M, Götte M, Melnyk RA. Auger A, et al. J Biol Chem. 2011 Aug 26;286(34):29575-83. doi: 10.1074/jbc.M111.268235. Epub 2011 Jul 7. J Biol Chem. 2011. PMID: 21737446 Free PMC article. - HIV-1 reverse transcriptase (RT) polymorphism 172K suppresses the effect of clinically relevant drug resistance mutations to both nucleoside and non-nucleoside RT inhibitors.
Hachiya A, Marchand B, Kirby KA, Michailidis E, Tu X, Palczewski K, Ong YT, Li Z, Griffin DT, Schuckmann MM, Tanuma J, Oka S, Singh K, Kodama EN, Sarafianos SG. Hachiya A, et al. J Biol Chem. 2012 Aug 24;287(35):29988-99. doi: 10.1074/jbc.M112.351551. Epub 2012 Jul 2. J Biol Chem. 2012. PMID: 22761416 Free PMC article. - Rate-limiting Pyrophosphate Release by HIV Reverse Transcriptase Improves Fidelity.
Li A, Gong S, Johnson KA. Li A, et al. J Biol Chem. 2016 Dec 16;291(51):26554-26565. doi: 10.1074/jbc.M116.753152. Epub 2016 Oct 24. J Biol Chem. 2016. PMID: 27777304 Free PMC article.
References
- Arion D., Kaushik,N., McCormick,S., Borkow,G. and Parniak,M.A. (1998) Phenotypic mechanism of HIV-1 resistance to 3′-azido-3′-deoxythymidine (AZT): increased polymerization processivity and enhanced sensitivity to pyrophosphate of the mutant viral reverse transcriptase. Biochemistry, 37, 15908–15917. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases