CENP-B box is required for de novo centromere chromatin assembly on human alphoid DNA - PubMed (original) (raw)
CENP-B box is required for de novo centromere chromatin assembly on human alphoid DNA
Jun-ichirou Ohzeki et al. J Cell Biol. 2002.
Abstract
Centromere protein (CENP) B boxes, recognition sequences of CENP-B, appear at regular intervals in human centromeric alpha-satellite DNA (alphoid DNA). In this study, to determine whether information carried by the primary sequence of alphoid DNA is involved in assembly of functional human centromeres, we created four kinds of synthetic repetitive sequences: modified alphoid DNA with point mutations in all CENP-B boxes, resulting in loss of all CENP-B binding activity; unmodified alphoid DNA containing functional CENP-B boxes; and nonalphoid repetitive DNA sequences with or without functional CENP-B boxes. These four synthetic repetitive DNAs were introduced into cultured human cells (HT1080), and de novo centromere assembly was assessed using the mammalian artificial chromosome (MAC) formation assay. We found that both the CENP-B box and the alphoid DNA sequence are required for de novo MAC formation and assembly of functional centromere components such as CENP-A, CENP-C, and CENP-E. Using the chromatin immunoprecipitation assay, we found that direct assembly of CENP-A and CENP-B in cells with synthetic alphoid DNA required functional CENP-B boxes. To the best of our knowledge, this is the first reported evidence of a functional molecular link between a centromere-specific DNA sequence and centromeric chromatin assembly in humans.
Figures
Figure 1.
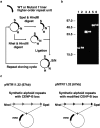
Configuration of alphoid arrays of the chromosome 21 centromere, and construction of the synthetic alphoid repeat unit with modified CENP-B boxes (mutant α21-I 11mer). (a) Capital letters E and X indicate EcoRI and XbaI cutting sites, respectively. The numeric characters below the α21-I 11mer unit indicate monomer numbers. (b) The 17-bp motif of the CENP-B box and its position in α21-I 11mer monomer units. The nucleotide sequence shaded black is necessary for CENP-B binding. The 17-bp sequence motif of monomer 10 has two nucleotide variations, and lacks CENP-B binding activity. All five CENP-B box motifs in the mutant α21-I 11mer were identical to this monomer 10 motif.
Figure 2.
Gel mobility-shift competition analysis of wild-type and mutant α21-I 11mer subunits. (a) α32P-CTP–labeled oligonucleotide probe (CB59) containing a CENP-B box sequence was mixed with HeLa nuclear extracts. The mobility shift of the probe with the CENP-B protein dimer complex, indicated by the arrow point (lane Ex), was blocked by preincubation with anti–CENP-B antibodies (lane Ab). Unlabeled PCR products of wild-type and mutant 2-, 4-, and 5mer subunits were used as competitors for the labeled CB59 probe with ×0, ×1, and ×2 and ×1, ×10, and ×100 excess of molecules, respectively. Capital letters indicate no extract (NE), probe with extract (Ex), and probe with extract preincubated with anti–CENP-B antibody (Ab). (b) Relative intensities of shifted bands are plotted in these panels.
Figure 3.
Construction of synthetic alphoid repeats with wild-type and mutant α21-I 11mer repeat units. (a) Schematic diagram of tandem concatenation of wild-type and mutant α21-I 11mer repeat units. (b) Pulse field gel electrophoresis of tandemly concatenated α21-I 11mer repeat arrays. Lanes 1, 3, and 5, endonuclease HindIII-cut BAC plasmids containing 8, 16, and 32 copies of extended wild type α21-I 11mer repeat units, respectively. Lanes 2, 4, and 6, BAC plasmids containing 8, 16, and 32 copies of extended mutant α21-I 11mer repeat units, respectively. (c) Circular BAC constructs of pWTR11.32 and pMTR11.32 have a common vector sequence containing a neomycin resistance gene. The only differences between the two plasmids are two nucleotide substitutions at each CENP-B box.
Figure 4.
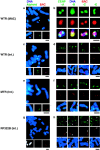
Examples of extrachromosomal events and centromere protein assembly on introduced DNAs. In all panels, DNA was counterstained with DAPI (blue), and introduced BAC DNA was detected by FISH with a fluorescence-labeled BAC probe (red). (a, c, e, and g) Chromosomal events detected by FISH. Green signals indicate α21-I 11mer DNA probe. (b, d, f, and h) Centromere protein assembly on the BAC DNA was analyzed with indirect immunofluorescent staining combined with FISH. Green signals indicate the indirect immunofluorescence of centromere proteins (CENP-A, -B, -C, and –E). In panel h, white arrowheads indicate the integration site of pRF322B.192. The following data from cell lines were used as examples: W0210R-8 (a and b), W0210R-1 (c and d), M1319 (e and f), and B0106 (g and h). Bars: (a and c–h) 5 μm; (b) 1 μm.
Figure 5.
CHIP analysis of centromere protein assembly on introduced alphoid DNA. (a) pWTR11.32-transformed cells (W0203 or W0210R-1) were mixed with reference pMTR11.32-transformed cells (M1319). (b) EcoRV-digested PCR products from genomic DNA templates. W0203 and M1319 cells were mixed at different ratios of cell number (from 16:1 to 1:16), and genomic DNAs for competitive PCR were prepared from these mixtures. Gray arrowheads indicate the 142-bp PCR fragment from WTR. Black arrowheads indicate the 111-bp PCR fragment from MTR. (c) EcoRV- digested PCR products from immunoprecipitated DNA template. Left four lanes show PCR products from diluted input DNA at 10−1, 10−2, 10−3, and 10−4 dilution, from left to right. PCR products from immunoprecipitated DNAs are shown in the right four lanes. Capital letters indicate antibodies used in immunoprecipitation: normal mouse IgG (IgG), anti–CENP-A (A), anti–CENP-B (B), and anti–histone H3 (H3). Two different rates of amplification, logarithmic amplification state (25–30 cycles) and plateau state (55 cycles), are shown in the top and bottom panels, respectively. DNA samples comprised 75% and 15% of total PCR products in the top and bottom panels, respectively. Relative concentrations of the WTR DNA fragment versus the MTR DNA fragment, as indicated by CHIP assay using each antibody, are shown below the panels.
Figure 6.
Construction of nonalphoid sequences with or without CENP-B boxes and CHIP analysis. (a) Schematic diagram of the construction of nonalphoid repetitive sequences containing CENP-B boxes (RF322B) and not containing CENP-B boxes (RF322L). pBR322-derived fragment (RF322) was ligated to the CENP-B box (RF322B) or spacer oligonucleotide sequence (RF322L). The resulting fragments were tandemly concatenated using the same method used to construct the α21-I 11mer alphoid repeat unit. (b) Competitive CHIP analysis of a pRF322B.192- transformed cell (B0106) using a pRF322L-transformed cell (L0102) as reference. Using the specific primer set, both synthetic RF322 fragments were amplified with competitive PCR, as in Fig. 5. The PCR products were separated by agarose gel electrophoresis after endonuclease EcoRI treatment. The ratio of concentration of the RF322B fragment to that of the RF322L fragment was calculated and is indicated below the panels.
Similar articles
- Structural rearrangements and insertions of dispersed elements in pericentromeric alpha satellites occur preferably at kinkable DNA sites.
Mashkova TD, Oparina NY, Lacroix MH, Fedorova LI, G Tumeneva I, Zinovieva OL, Kisselev LL. Mashkova TD, et al. J Mol Biol. 2001 Jan 5;305(1):33-48. doi: 10.1006/jmbi.2000.4270. J Mol Biol. 2001. PMID: 11114245 - A minimal CENP-A core is required for nucleation and maintenance of a functional human centromere.
Okamoto Y, Nakano M, Ohzeki J, Larionov V, Masumoto H. Okamoto Y, et al. EMBO J. 2007 Mar 7;26(5):1279-91. doi: 10.1038/sj.emboj.7601584. Epub 2007 Feb 22. EMBO J. 2007. PMID: 17318187 Free PMC article. - Epigenetic assembly of centromeric chromatin at ectopic alpha-satellite sites on human chromosomes.
Nakano M, Okamoto Y, Ohzeki J, Masumoto H. Nakano M, et al. J Cell Sci. 2003 Oct 1;116(Pt 19):4021-34. doi: 10.1242/jcs.00697. J Cell Sci. 2003. PMID: 12953060 - The role of CENP-B and alpha-satellite DNA: de novo assembly and epigenetic maintenance of human centromeres.
Masumoto H, Nakano M, Ohzeki J. Masumoto H, et al. Chromosome Res. 2004;12(6):543-56. doi: 10.1023/B:CHRO.0000036593.72788.99. Chromosome Res. 2004. PMID: 15289662 Review. - Human artificial chromosome: Chromatin assembly mechanisms and CENP-B.
Ohzeki JI, Otake K, Masumoto H. Ohzeki JI, et al. Exp Cell Res. 2020 Apr 15;389(2):111900. doi: 10.1016/j.yexcr.2020.111900. Epub 2020 Feb 8. Exp Cell Res. 2020. PMID: 32044309 Review.
Cited by
- CENP-A: the key player behind centromere identity, propagation, and kinetochore assembly.
De Rop V, Padeganeh A, Maddox PS. De Rop V, et al. Chromosoma. 2012 Dec;121(6):527-38. doi: 10.1007/s00412-012-0386-5. Epub 2012 Oct 26. Chromosoma. 2012. PMID: 23095988 Free PMC article. Review. - DNA Sequence-Specific Binding of CENP-B Enhances the Fidelity of Human Centromere Function.
Fachinetti D, Han JS, McMahon MA, Ly P, Abdullah A, Wong AJ, Cleveland DW. Fachinetti D, et al. Dev Cell. 2015 May 4;33(3):314-27. doi: 10.1016/j.devcel.2015.03.020. Dev Cell. 2015. PMID: 25942623 Free PMC article. - Sequences associated with centromere competency in the human genome.
Hayden KE, Strome ED, Merrett SL, Lee HR, Rudd MK, Willard HF. Hayden KE, et al. Mol Cell Biol. 2013 Feb;33(4):763-72. doi: 10.1128/MCB.01198-12. Epub 2012 Dec 10. Mol Cell Biol. 2013. PMID: 23230266 Free PMC article. - Population and subspecies diversity at mouse centromere satellites.
Arora UP, Charlebois C, Lawal RA, Dumont BL. Arora UP, et al. BMC Genomics. 2021 Apr 17;22(1):279. doi: 10.1186/s12864-021-07591-5. BMC Genomics. 2021. PMID: 33865332 Free PMC article. - A novel cell response triggered by interphase centromere structural instability.
Morency E, Sabra M, Catez F, Texier P, Lomonte P. Morency E, et al. J Cell Biol. 2007 Jun 4;177(5):757-68. doi: 10.1083/jcb.200612107. J Cell Biol. 2007. PMID: 17548509 Free PMC article.
References
- Abrieu, A., J.A. Kahana, K.W. Wood, and D.W. Cleveland. 2000. CENP-E as an essential component of the mitotic checkpoint in vitro. Cell. 102:817–826. - PubMed
- Alexandrov, I., A. Kazakov, I. Tumeneva, V. Shepelev, and Y. Yurov. 2001. α-Satellite DNA of primates: old and new families. Chromosoma. 110:253–266. - PubMed
- Choo, K.H.A. 1997a. The Centromere. Oxford University Press, Oxford, UK. 320 pp.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources