Restoration of LDL receptor function in cells from patients with autosomal recessive hypercholesterolemia by retroviral expression of ARH1 - PubMed (original) (raw)
Restoration of LDL receptor function in cells from patients with autosomal recessive hypercholesterolemia by retroviral expression of ARH1
Emily R Eden et al. J Clin Invest. 2002 Dec.
Abstract
Familial hypercholesterolemia is an autosomal dominant disorder with a gene-dosage effect that is usually caused by mutations in the LDL receptor gene that disrupt normal clearance of LDL. In the homozygous form, it results in a distinctive clinical phenotype, characterized by inherited hypercholesterolemia, cholesterol deposition in tendons, and severe premature coronary disease. We described previously two families with autosomal recessive hypercholesterolemia that is not due to mutations in the LDL receptor gene but is characterized by defective LDL receptor-dependent internalization and degradation of LDL by transformed lymphocytes from the patients. We mapped the defective gene to chromosome 1p36 and now show that the disorder in these and a third English family is due to novel mutations in ARH1, a newly identified gene encoding an adaptor-like protein. Cultured skin fibroblasts from affected individuals exhibit normal LDL receptor activity, but their monocyte-derived macrophages are similar to transformed lymphocytes, being unable to internalize and degrade LDL. Retroviral expression of normal human ARH1 restores LDL receptor internalization in transformed lymphocytes from an affected individual, as demonstrated by uptake and degradation of (125)I-labeled LDL and confocal microscopy of cells labeled with anti-LDL-receptor Ab.
Figures
Figure 1
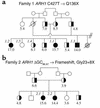
Pedigrees of family 1 (a) and family 2 (b). The plasma cholesterol concentration (mmol/l) is shown below each symbol. Filled symbols, homozygous for the Q136X (a, family 1) or the delGG86,87 (b, family 2) mutations in ARH1; half-filled symbols, heterozygous carriers of the mutation, confirmed by sequencing amplified fragments of genomic DNA. Individuals whose cells were analyzed for LDL receptor activity or ARH1 mRNA levels are identified by a number in italics.
Figure 2
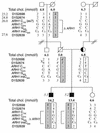
Pedigree and haplotype analysis of family 3. The plasma cholesterol (chol.) concentration (mmol/l) is shown below each symbol. Filled symbols, homozygous for the insC mutation in ARH1; half-filled symbols, heterozygous carriers of the mutation, confirmed by sequencing amplified fragments of genomic DNA. Haplotypes for the ARH1 locus are shown for polymorphic markers flanking the gene, known polymorphisms (8), and the two novel base substitutions within the coding region of ARH1; the position of each marker on Ensembl is shown (Mb). A recombination in the paternal uncle of the proband is indicated (X). The maternal allele carrying the insC620 mutation is boxed; the paternal allele carrying the putative deletion is boxed and shaded. The haplotype in the father, shown in italics, was deduced. Individuals whose cells were analyzed for LDL receptor activity or ARH1 mRNA levels are identified by a number in italics.
Figure 3
Identification of a partial deletion of ARH1 in family 3. (a) Diagram showing the approximate size of the ARH1 DNA probes used for fluorescent in situ hybridization (FISH). The vertical black bars represent the nine exons of ARH1. (b) Metaphase chromosomes from individual FH3.1 were hybridized with either DIG-labeled ARH1 DNA probe 1 or probe 2 as indicated (detected with FITC-labeled anti-DIG Ab; small filled arrows) and biotin-labeled chromosome 1–specific α-satellite probe DNA (detected with Cy3-labeled anti-biotin Ab; large open arrows).
Figure 4
Degradation of 125I-labeled LDL by cultured skin fibroblasts and EBV- lymphocytes from individuals in family 1. Cells were preincubated for 16 hours in medium containing LPDS and then for 4 hours with 125I-labeled LDL. Saturable degradation of LDL was determined as the difference in the amount of trichloroacetic acid–soluble (TCA-soluble), non-iodide radioactivity in the medium of cells incubated in the presence and absence of an excess of unlabeled LDL (1 mg/ml); values are the mean of duplicate dishes. Nonsaturable degradation of LDL by normal cells was always less than 5% of the total. Data shown are representative of at least two separate experiments. (a) Cultured skin fibroblasts from the proband in family 1 (ARH–/–), and from a normolipemic control (ARH+/+). (b) EBV-lymphocytes from the proband in family 1 (ARH–/–), her heterozygous sibling (ARH+/–), and a sibling who does not carry the mutant allele (ARH+/+).
Figure 5
Degradation of 125I-labeled LDL by skin fibroblasts, EBV-lymphocytes, and monocyte-derived macrophages from individuals in family 3. Cells were preincubated for 16 hours in medium containing LPDS and then for 4 hours with 125I-labeled LDL. Saturable degradation of LDL was determined as the difference in the amount of TCA-soluble, non-iodide radioactivity in the medium of cells incubated in the presence and absence of an excess of unlabeled LDL (1 mg/ml); values are the mean of duplicate dishes. Nonsaturable degradation of LDL by normal cells was always less than 5% of the total. Data shown are representative of at least two separate experiments. (a) EBV-lymphocytes, (b) skin fibroblasts, and (c) monocyte-derived macrophages, from probands 3.1 (filled triangles) and 3.2 (open triangles) in family 3 and from unrelated normolipemic controls (filled circles).
Figure 6
Effect of expression of c-myc-ARH1 on LDL receptor activity in mutant EBV-lymphocytes. (a and b) Cells were preincubated for 16 hours with LPDS before preparation of cell extracts. Proteins were fractionated on nonreduced SDS-polyacrylamide gels (13%), transferred to nylon membranes, and immunoblotted with anti–c-myc (lanes 1–8) or anti–LDL receptor Ab (lanes 9 and 10). Bound Ab was detected with peroxidase-conjugated anti-mouse IgG and chemiluminescence. (a) Whole-cell extracts (approximately 50 μg of protein per lane) of Chinese hamster ovary (CHO) cells transiently transfected with pcDNA3-c-myc-ARH1PA317 (lane 1), PA317 cells transfected with ARH1 retroviral construct (lane 2), PA317 cells (lane 3), EBV-lymphocytes from affected individual 1.1 (ARH1.1 cells) 1 month after infection with c-myc-ARH1 retrovirus (lane 4), and uninfected EBV-lymphocytes from affected individual 1.1 (lane 5). (b) Whole-cell extracts (approximately 50 μg of protein per lane) of EBV-lymphocytes from affected individual 1.1 (lane 6), the same cells 3 months after stable infection with c-myc-ARH1 retrovirus (lanes 7 and 9), and the same infected cells after preincubation for 16 hours with 0.3 μM trichostatin A (lane 8 and 10). (c) Virus-infected (ARH–/c-myc-ARH) and uninfected (ARH–) EBV-lymphocytes from individual 1.1 were preincubated for 16 hours in medium containing LPDS and then for 4 hours with 125I-labeled LDL. Saturable degradation of LDL was determined as the difference in the amount of TCA-soluble, non-iodide radioactivity in the medium of cells incubated in the presence and absence of an excess of unlabeled LDL (1 mg/ml); values are the mean of duplicate dishes. Data shown are representative of two separate experiments.
Figure 7
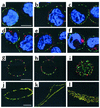
Confocal microscopy of cells labeled with anti–LDL receptor Ab. (a–f) EBV-lymphocytes were incubated with rabbit anti–LDL receptor (red) at 4°C, washed at 4°C, and either directly permeabilized (a–c) or incubated for 10 minutes at 37°C before permeabilization to allow internalization of LDL receptor/Ab complexes (d–f). Permeabilized cells were then incubated with mouse anti–α-adaptin (AP2), washed, and incubated with Alexa 568–conjugated goat anti-rabbit IgG (LDL receptors, red) and Alexa 488–conjugated goat anti-mouse IgG (AP2, green). Nuclei were stained with DAPI. The plates shown are an overlay of red and green images. The bar represents 5.0 μm. (a and d) Control cells; (b and e) cells from proband 1.1; (c and f) cells from proband 1.1 expressing viral c-myc-ARH. (g–l) EBV-lymphocytes (g–i) or cultured skin fibroblasts (j–l) from three different control subjects were incubated with anti–LDL receptor Ab at 4°C, permeabilized, and then incubated with anti–α-adaptin Ab (AP2) as described for a–f above. The bars represent 5.0 μm (in g for g–i, and in j for j–l).
Similar articles
- Use of homozygosity mapping to identify a region on chromosome 1 bearing a defective gene that causes autosomal recessive homozygous hypercholesterolemia in two unrelated families.
Eden ER, Naoumova RP, Burden JJ, McCarthy MI, Soutar AK. Eden ER, et al. Am J Hum Genet. 2001 Mar;68(3):653-60. doi: 10.1086/318795. Epub 2001 Feb 9. Am J Hum Genet. 2001. PMID: 11179013 Free PMC article. - Normal sorting but defective endocytosis of the low density lipoprotein receptor in mice with autosomal recessive hypercholesterolemia.
Jones C, Hammer RE, Li WP, Cohen JC, Hobbs HH, Herz J. Jones C, et al. J Biol Chem. 2003 Aug 1;278(31):29024-30. doi: 10.1074/jbc.M304855200. Epub 2003 May 13. J Biol Chem. 2003. PMID: 12746448 - A combined LDL receptor/LDL receptor adaptor protein 1 mutation as the cause for severe familial hypercholesterolemia.
Soufi M, Rust S, Walter M, Schaefer JR. Soufi M, et al. Gene. 2013 May 25;521(1):200-3. doi: 10.1016/j.gene.2013.03.034. Epub 2013 Mar 17. Gene. 2013. PMID: 23510778 - Autosomal recessive hypercholesterolemia.
Soutar AK, Naoumova RP. Soutar AK, et al. Semin Vasc Med. 2004 Aug;4(3):241-8. doi: 10.1055/s-2004-861491. Semin Vasc Med. 2004. PMID: 15630633 Review. - The molecular genetic basis and diagnosis of familial hypercholesterolemia in Denmark.
Jensen HK. Jensen HK. Dan Med Bull. 2002 Nov;49(4):318-45. Dan Med Bull. 2002. PMID: 12553167 Review.
Cited by
- The AP-2 adaptor beta2 appendage scaffolds alternate cargo endocytosis.
Keyel PA, Thieman JR, Roth R, Erkan E, Everett ET, Watkins SC, Heuser JE, Traub LM. Keyel PA, et al. Mol Biol Cell. 2008 Dec;19(12):5309-26. doi: 10.1091/mbc.e08-07-0712. Epub 2008 Oct 8. Mol Biol Cell. 2008. PMID: 18843039 Free PMC article. - Molecular characterization of proprotein convertase subtilisin/kexin type 9-mediated degradation of the LDLR.
Wang Y, Huang Y, Hobbs HH, Cohen JC. Wang Y, et al. J Lipid Res. 2012 Sep;53(9):1932-43. doi: 10.1194/jlr.M028563. Epub 2012 Jul 4. J Lipid Res. 2012. PMID: 22764087 Free PMC article. - Caenorhabditis elegans reveals a FxNPxY-independent low-density lipoprotein receptor internalization mechanism mediated by epsin1.
Kang YL, Yochem J, Bell L, Sorensen EB, Chen L, Conner SD. Kang YL, et al. Mol Biol Cell. 2013 Feb;24(3):308-18. doi: 10.1091/mbc.E12-02-0163. Epub 2012 Dec 14. Mol Biol Cell. 2013. PMID: 23242996 Free PMC article. - Sorting it out: AP-2 and alternate clathrin adaptors in endocytic cargo selection.
Traub LM. Traub LM. J Cell Biol. 2003 Oct 27;163(2):203-8. doi: 10.1083/jcb.200309175. J Cell Biol. 2003. PMID: 14581447 Free PMC article. Review. - Monogenic hypercholesterolemia: new insights in pathogenesis and treatment.
Rader DJ, Cohen J, Hobbs HH. Rader DJ, et al. J Clin Invest. 2003 Jun;111(12):1795-803. doi: 10.1172/JCI18925. J Clin Invest. 2003. PMID: 12813012 Free PMC article. Review. No abstract available.
References
- Goldstein, J.L., Hobbs, H., and Brown, M.S. 1995. Familial hypercholesterolemia. In The metabolic and molecular bases of inherited disease. C.R. Scriver, A.L. Beaudet, W.S. Sly, and D. Valle, editors. McGraw-Hill. New York, New York, USA. 1981–2030.
- Garcia CK, et al. Autosomal recessive hypercholesterolemia caused by mutations in a putative LDL receptor adaptor protein. Science. 2001;292:1394–1398. - PubMed
- Zuliani G, et al. Characterization of a new form of inherited hypercholesterolemia: familial recessive hypercholesterolemia. Arterioscler Thromb Vasc Biol. 1999;19:802–809. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases