Antisense inhibition of gene expression in cells by oligonucleotides incorporating locked nucleic acids: effect of mRNA target sequence and chimera design - PubMed (original) (raw)
Antisense inhibition of gene expression in cells by oligonucleotides incorporating locked nucleic acids: effect of mRNA target sequence and chimera design
Dwaine A Braasch et al. Nucleic Acids Res. 2002.
Abstract
Use of antisense oligonucleotides is a versatile strategy for achieving control of gene expression. Unfortunately, the interpretation of antisense-induced phenotypes is sometimes difficult, and chemical modifications that improve the potency and specificity of antisense action would be useful. The introduction of locked nucleic acid (LNA) bases into oligonucleotides confers exceptional improvement in binding affinity, up to 10 degrees C per substitution, making LNAs an exciting option for the optimization of antisense efficacy. Here we examine the rules governing antisense gene inhibition within cells by oligonucleotides that contain LNA bases. LNA- containing oligomers were transfected into cells using cationic lipid and accumulated in the nucleus. We tested antisense gene inhibition by LNAs and LNA-DNA chimeras complementary to the 5'-untranslated region, the region surrounding the start codon and the coding region of mRNA, and identified effective antisense agents targeted to each of these locations. Our data suggest that LNA bases can be used to develop antisense oligonucleotides and that their use is a versatile approach for efficiently inhibiting gene expression inside cells.
Figures
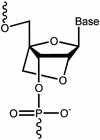
Figure 1
Structure of locked nucleic acid (LNA).
Figure 2
Schematic of luciferase mRNA showing the approximate target sites of LNA and LNA–DNA chimeras used in these studies.
Figure 3
Images of CV-1 cells transfected with a LNA–DNA chimera that was analogous in sequence to XV and labeled with Cy3 fluorophore. All images are magnified 630 times and the field of view is the same. (Left to right) DIC bright field image; staining with the cell permeable nuclear dye Hoechst 32358; localization of Cy3-labeled LNA–DNA chimera; overlay of image of Hoechst 33258 and Cy3-labeled LNA–DNA chimera.
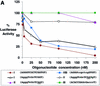
Figure 4
Dose–response curves of antisense gene inhibition by LNAs and LNA–DNA chimeras targeted to (A) the 5′-UTR, (B) the translation start site and (C) the downstream coding region of luciferase mRNA. All points are averages of triplicate determinations and are normalized to an independent measurement of β-galactosidase activity. Capitalized letters within a sequence represent LNA, lower case letters represent DNA bases and underlined bases depict mismatched bases.
Figure 4
Dose–response curves of antisense gene inhibition by LNAs and LNA–DNA chimeras targeted to (A) the 5′-UTR, (B) the translation start site and (C) the downstream coding region of luciferase mRNA. All points are averages of triplicate determinations and are normalized to an independent measurement of β-galactosidase activity. Capitalized letters within a sequence represent LNA, lower case letters represent DNA bases and underlined bases depict mismatched bases.
Figure 4
Dose–response curves of antisense gene inhibition by LNAs and LNA–DNA chimeras targeted to (A) the 5′-UTR, (B) the translation start site and (C) the downstream coding region of luciferase mRNA. All points are averages of triplicate determinations and are normalized to an independent measurement of β-galactosidase activity. Capitalized letters within a sequence represent LNA, lower case letters represent DNA bases and underlined bases depict mismatched bases.
Similar articles
- Inhibition of gene expression inside cells by peptide nucleic acids: effect of mRNA target sequence, mismatched bases, and PNA length.
Doyle DF, Braasch DA, Simmons CG, Janowski BA, Corey DR. Doyle DF, et al. Biochemistry. 2001 Jan 9;40(1):53-64. doi: 10.1021/bi0020630. Biochemistry. 2001. PMID: 11141056 - Intracellular inhibition of hepatitis C virus (HCV) internal ribosomal entry site (IRES)-dependent translation by peptide nucleic acids (PNAs) and locked nucleic acids (LNAs).
Nulf CJ, Corey D. Nulf CJ, et al. Nucleic Acids Res. 2004 Jul 19;32(13):3792-8. doi: 10.1093/nar/gkh706. Print 2004. Nucleic Acids Res. 2004. PMID: 15263060 Free PMC article. - A novel dual lock method for down-regulation of genes, in which a target mRNA is captured at 2 independent positions by linked locked nucleic acid antisense oligonucleotides.
Takata R, Makado G, Kitamura A, Watanabe H, Wada T. Takata R, et al. RNA Biol. 2016;13(3):279-89. doi: 10.1080/15476286.2015.1119364. Epub 2016 Feb 18. RNA Biol. 2016. PMID: 26890856 Free PMC article. - Locked nucleic acids: a promising molecular family for gene-function analysis and antisense drug development.
Orum H, Wengel J. Orum H, et al. Curr Opin Mol Ther. 2001 Jun;3(3):239-43. Curr Opin Mol Ther. 2001. PMID: 11497347 Review. - Locked nucleic acid oligonucleotides: the next generation of antisense agents?
Grünweller A, Hartmann RK. Grünweller A, et al. BioDrugs. 2007;21(4):235-43. doi: 10.2165/00063030-200721040-00004. BioDrugs. 2007. PMID: 17628121 Review.
Cited by
- Comparison of different antisense strategies in mammalian cells using locked nucleic acids, 2'-O-methyl RNA, phosphorothioates and small interfering RNA.
Grünweller A, Wyszko E, Bieber B, Jahnel R, Erdmann VA, Kurreck J. Grünweller A, et al. Nucleic Acids Res. 2003 Jun 15;31(12):3185-93. doi: 10.1093/nar/gkg409. Nucleic Acids Res. 2003. PMID: 12799446 Free PMC article. - Enhancing Antisense Oligonucleotide-Based Therapeutic Delivery with DG9, a Versatile Cell-Penetrating Peptide.
Haque US, Yokota T. Haque US, et al. Cells. 2023 Oct 2;12(19):2395. doi: 10.3390/cells12192395. Cells. 2023. PMID: 37830609 Free PMC article. Review. - Selection, optimization, and pharmacokinetic properties of a novel, potent antiviral locked nucleic acid-based antisense oligomer targeting hepatitis C virus internal ribosome entry site.
Laxton C, Brady K, Moschos S, Turnpenny P, Rawal J, Pryde DC, Sidders B, Corbau R, Pickford C, Murray EJ. Laxton C, et al. Antimicrob Agents Chemother. 2011 Jul;55(7):3105-14. doi: 10.1128/AAC.00222-11. Epub 2011 Apr 18. Antimicrob Agents Chemother. 2011. PMID: 21502629 Free PMC article. - Delivery of RNAi-Based Oligonucleotides by Electropermeabilization.
Chabot S, Pelofy S, Teissié J, Golzio M. Chabot S, et al. Pharmaceuticals (Basel). 2013 Apr 10;6(4):510-21. doi: 10.3390/ph6040510. Pharmaceuticals (Basel). 2013. PMID: 24276121 Free PMC article. - Use of steric blocking antisense oligonucleotides for the targeted inhibition of junction containing precursor microRNAs.
Ma S, Howden SA, Keane SC. Ma S, et al. bioRxiv [Preprint]. 2024 Apr 8:2024.04.08.588531. doi: 10.1101/2024.04.08.588531. bioRxiv. 2024. PMID: 38645194 Free PMC article. Preprint.
References
- Braasch D.A. and Corey,D.R. (2002) Novel antisense strategies for controlling gene expression. Biochemistry, 41, 4503–4510. - PubMed
- Koshkin A.A., Singh,S.K., Nielsen,P., Rajwanshi,V.K., Kumar,R., Meldgaard,M., Olsen,C.E. and Wengel,J. (1998) LNA (locked nucleic acid): synthesis of the adenine, cytosine, guanine, 5-methyl cytosine, thymine, and uracil bicyclonucleoside monomers, oligomerisation and unprecedented nucleic acid recognition. Tetrahedron, 54, 3607–3630.
- Obika S., Nanbu,D., Hari,Y., Andoh,J., Morio,K., Doi,T. and Imanishi,T. (1998) Stability and structural features of the duplexes containing nucleoside analogues with fixed N-type conformation. Tetrahedron Lett., 39, 5401–5404.
- Wengel J. (1999) Synthesis of 3′-C- and 4′-C-branched oligodeoxynucleotides and the development of locked nucleic acid (LNA). Acc. Chem. Res., 32, 301–310.
- Braasch D.A. and Corey,D.R. (2001) Locked nucleic acid (LNA): fine-tuning the recognition of DNA and RNA. Chem. Biol., 8, 1–7. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources