A subset of chaperones and folding enzymes form multiprotein complexes in endoplasmic reticulum to bind nascent proteins - PubMed (original) (raw)
A subset of chaperones and folding enzymes form multiprotein complexes in endoplasmic reticulum to bind nascent proteins
Laurent Meunier et al. Mol Biol Cell. 2002 Dec.
Abstract
We demonstrate the existence of a large endoplasmic reticulum (ER)-localized multiprotein complex that is comprised of the molecular chaperones BiP; GRP94; CaBP1; protein disulfide isomerase (PDI); ERdj3, a recently identified ER Hsp40 cochaperone; cyclophilin B; ERp72; GRP170; UDP-glucosyltransferase; and SDF2-L1. This complex is associated with unassembled, incompletely folded immunoglobulin heavy chains. Except for ERdj3, and to a lesser extent PDI, this complex also forms in the absence of nascent protein synthesis and is found in a variety of cell types. Cross-linking studies reveal that the majority of these chaperones are included in the complex. Our data suggest that this subset of ER chaperones forms an ER network that can bind to unfolded protein substrates instead of existing as free pools that assembled onto substrate proteins. It is noticeable that most of the components of the calnexin/calreticulin system, which include some of the most abundant chaperones inside the ER, are either not detected in this complex or only very poorly represented. This study demonstrates an organization of ER chaperones and folding enzymes that has not been previously appreciated and suggests a spatial separation of the two chaperone systems that may account for the temporal interactions observed in other studies.
Figures
Figure 1
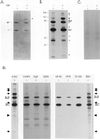
Unassembled Ig heavy chains associate with a number of ER proteins in addition to BiP. The postnuclear fraction was obtained from metabolically labeled Ag8(8) cells and divided into two tubes: one for a negative control and the other treated with 150 μg/ml DSP. After quenching, vesicles were solubilized and heavy chains were precipitated with protein A-Sepharose for SDS-PAGE analysis under reducing conditions. The gel was first stained with Coomassie Blue (A) and then exposed to film for an autoradiograph (B). BiP and a γ heavy chain are indicated on the left of the gel. Additional proteins detected after cross-linking are marked with arrows. Ag8.653 cells were labeled and treated similarly (− and +DSP) and then immunoprecipitated with protein A-Sepharose (C). The cell lysates were obtained by using different detergents to release the ER proteins (1% CHAPS, 1% digitonin, 1% dodecylmaltoside [DDM], 0.5% deoxycholic acid, 0.5% NP-40, and 0.2% Triton X-100) or no detergent (a cycle of freeze-thawing followed by homogenization of the postnuclear fraction in HFTB buffer (HFTB) or 10-s sonication in TESV buffer (TESV) as described in MATERIALS AND METHODS (D). Additional proteins detected after cross-linking and after sonication are indicated with arrows.
Figure 2
Heavy chain–BiP complexes contain substantial quantities of GRP94 and smaller amounts of other chaperones and folding enzymes. Total cell lysates from 1 × 106 Ag8(8) and Ag8.653 cells were used as a positive control for the antibodies (lanes 1 and 4). The postnuclear fraction from 10 × 106 cells was either treated with DSP (lanes 3 and 6) or left untreated (lanes 2 and 5) and lysates were prepared as described in MATERIALS AND METHODS. Lysates from Ag8(8) (lanes 2 and 3) and from Ag8.653 (lanes 5 and 6) were incubated with protein A-Sepharose and precipitated proteins were electrophoresed and transferred for blotting. Segments of the nitrocellulose were reacted with the antibodies indicated and developed as described (A). Then 10 × 106 Ag8(8) cells were treated overnight with 2 (+) or 0 (−) μg/ml tunicamycin, incubated with DSP, and heavy chains were isolated from lysates with protein A-Sepharose and processed as in A. Total cell lysate from 10 × 106 cells was used as a positive control for the antibodies (B). Cells were treated as in B except that 40 × 106 Ag8(8) cells per lane were used, and only 1/10 of the lysate was used as a positive control for the antibodies (C).
Figure 3
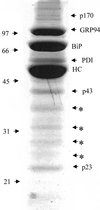
Coomassie staining of the proteins of the complex for isolation and identification. Then 300 × 106 Ag8(8) cells were treated with 150 μg/ml DSP. The postnuclear fraction was prepared, and Ig heavy chains and associated proteins were isolated with protein A-Sepharose, which were separated by reducing SDS-PAGE on a 10% acrylamide gel. The gel was then Coomassie stained and protein bands were isolated and submitted to mass spectrometry analysis. The arrows shows all the proteins analyzed and the asterisk (*) shows degradation products from the heavy chain.
Figure 4
Sequences of four other proteins of the complex. The following sequences were obtained from the p170, p43, and p23 bands associated with heavy chains as described in MATERIALS AND METHODS. The sequences of UDP-GT (A), the ER Hsp40 cochaperone-ERdj3 (B), cyclophilin B (C), and SDF2-L1 (D). The peptide sequences obtained for each protein are shown in bold.
Figure 5
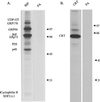
ER molecular chaperones exist as a complex in the absence of heavy chains. Ag8.653 cells were labeled for 16 h. The postnuclear fraction was isolated and treated with DSP as described in MATERIALS AND METHODS. Lysate was precipitated as indicated with anti-BiP antibody (A) or anti-calreticulin antibody (B). Protein A-Sepharose only was used as a control for nonspecific precipitation.
Figure 6
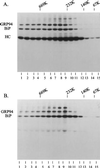
Additional ER chaperones bind to the same domain of heavy chain as BiP. Ag8(8) (lanes 1 and 2) and G403 (lanes 3–6) cells (heavy chain [HC] ΔCH1+, LC−) were labeled overnight, split in two aliquots, and treated with 0 or 100 μg/ml DSP. Gamma heavy chains were precipitated with protein A-Sepharose and analyzed by reducing SDS-PAGE. The right panel is the same thing that the middle panel but are from an autoradiograph that was exposed 15 times longer.
Figure 7
Sequence of CaBP1. Sequence of CaBP1 was obtained from 200 × 106 G403 cells. Cells were cross-linked, lysed in NP-40 lysing buffer, and precipitated with protein A-Sepharose. The precipitate was then resolved by reducing SDS-PAGE and the gel Coomassie stained. The sequence was obtained as described in MATERIALS AND METHODS.
Figure 8
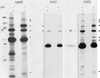
Multiprotein complex remains upon treatment with cycloheximide to inhibit the translation of new proteins. Ag8.653 cells (8 × 106) were left untreated or treated with 50 μg/ml cycloheximide for 2 h. The postnuclear fraction was prepared from cells, cross-linked with DSP, and lysed in NP-40 lysing buffer. The two cross-linked ER lysates were divided into five aliquots, each of which were directly applied to 5–15% gradient gels before transferring to nitrocellulose membranes for blotting. Untreated Ag8.653 cells (8 × 105) were lysed without prior cross-linking to identify the mobility of the free pool of the various proteins. The nitrocellulose membranes were reacted with the indicated polyclonal antisera. The mobility of the free pool of each protein is indicated with an arrow.
Figure 9
Visualization of the high-molecular-weight complexes by 2D gels. Ag8(8) or Ag8.653 cells (6 × 106) were metabolically labeled overnight and then treated with 150 μg/ml DSP. Protein complexes were immunoprecipitated with protein A-Sepharose alone for Ag8(8) or anti-BiP and protein A-Sepharose for Ag8.653. The samples were first electrophoresed under nonreducing conditions to separate different cross-linked complexes that might be present (1). The gel strip corresponding to a single sample was cut from the first gel and equilibrated in 5 ml of reducing SDS sample buffer for 40 min at room temperature on a rocker to reduce DSP and liberate the various proteins in the complex. The gel strip was then placed on the top of a second gel and run at a 90° angle to the first (2). After staining the gel was dried and a film exposed. A and B is obtained from Ag(8) and Ag8.653 cells, respectively.
Figure 10
Velocity gradient centrifugation to determine the size of the cross-linked complex. Ag8(8) and Ag8.653 cells (10 × 106) were metabolically labeled and the postnuclear fraction was prepared as described previously. Cross-linking was performed by including 150 μg/ml DSP in the HEPES buffer containing 0.25 M sucrose. Triton X-100 (1%) was added to the cross-linked sample and then the lysates and high-molecular-weight markers were centrifuged through 20–40% glycerol gradients. Each gradient was then separated into 15 fractions and the complex was precipitated with protein A-Sepharose beads for the Ag8(8) complexes (A) or immunoprecipitated with a polyclonal anti-BiP antibody for the Ag8.653 complexes (B).
Similar articles
- SDF2-like protein 1 (SDF2L1) regulates the endoplasmic reticulum localization and chaperone activity of ERdj3 protein.
Hanafusa K, Wada I, Hosokawa N. Hanafusa K, et al. J Biol Chem. 2019 Dec 13;294(50):19335-19348. doi: 10.1074/jbc.RA119.009603. Epub 2019 Oct 17. J Biol Chem. 2019. PMID: 31624144 Free PMC article. - Folding of thyroglobulin in the calnexin/calreticulin pathway and its alteration by loss of Ca2+ from the endoplasmic reticulum.
Di Jeso B, Ulianich L, Pacifico F, Leonardi A, Vito P, Consiglio E, Formisano S, Arvan P. Di Jeso B, et al. Biochem J. 2003 Mar 1;370(Pt 2):449-58. doi: 10.1042/BJ20021257. Biochem J. 2003. PMID: 12401114 Free PMC article. - Protein folding and quality control in the endoplasmic reticulum.
Kleizen B, Braakman I. Kleizen B, et al. Curr Opin Cell Biol. 2004 Aug;16(4):343-9. doi: 10.1016/j.ceb.2004.06.012. Curr Opin Cell Biol. 2004. PMID: 15261665 Review. - Calnexin cycle - structural features of the ER chaperone system.
Kozlov G, Gehring K. Kozlov G, et al. FEBS J. 2020 Oct;287(20):4322-4340. doi: 10.1111/febs.15330. Epub 2020 Apr 27. FEBS J. 2020. PMID: 32285592 Free PMC article. Review.
Cited by
- BRCA1 mediates protein homeostasis through the ubiquitination of PERK and IRE1.
Hromas R, Srinivasan G, Yang M, Jaiswal A, Totterdale TA, Phillips L, Kirby A, Khodayari N, Brantley M, Williamson EA, Kong KY. Hromas R, et al. iScience. 2022 Nov 19;25(12):105626. doi: 10.1016/j.isci.2022.105626. eCollection 2022 Dec 22. iScience. 2022. PMID: 36471805 Free PMC article. - KDEL Receptors: Pathophysiological Functions, Therapeutic Options, and Biotechnological Opportunities.
Cela I, Dufrusine B, Rossi C, Luini A, De Laurenzi V, Federici L, Sallese M. Cela I, et al. Biomedicines. 2022 May 25;10(6):1234. doi: 10.3390/biomedicines10061234. Biomedicines. 2022. PMID: 35740256 Free PMC article. Review. - The Impact of Glycoengineering on the Endoplasmic Reticulum Quality Control System in Yeasts.
Piirainen MA, Frey AD. Piirainen MA, et al. Front Mol Biosci. 2022 Jun 2;9:910709. doi: 10.3389/fmolb.2022.910709. eCollection 2022. Front Mol Biosci. 2022. PMID: 35720120 Free PMC article. Review. - Characterization of CNPY5 and its family members.
Schildknegt D, Lodder N, Pandey A, Chatsisvili A, Egmond M, Pena F, Braakman I, van der Sluijs P. Schildknegt D, et al. Protein Sci. 2019 Jul;28(7):1276-1289. doi: 10.1002/pro.3635. Epub 2019 May 16. Protein Sci. 2019. PMID: 31050855 Free PMC article. - Unfolded protein response and cell death after depletion of brefeldin A-inhibited guanine nucleotide-exchange protein GBF1.
Citterio C, Vichi A, Pacheco-Rodriguez G, Aponte AM, Moss J, Vaughan M. Citterio C, et al. Proc Natl Acad Sci U S A. 2008 Feb 26;105(8):2877-82. doi: 10.1073/pnas.0712224105. Epub 2008 Feb 14. Proc Natl Acad Sci U S A. 2008. PMID: 18287014 Free PMC article.
References
- Balow JP, Weissman JD, Kearse KP. Unique expression of major histocompatibility complex class I proteins in the absence of glucose trimming and calnexin association. J Biol Chem. 1995;270:29025–29029. - PubMed
- Bies C, Guth S, Janoschek K, Nastainczyk W, Volkmer J, Zimmermann R. A Scj1p homolog and folding catalysts present in dog pancreas microsomes. Biol Chem. 1999;380:1175–1182. - PubMed
- Blond-Elguindi S, Cwirla SE, Dower WJ, Lipshutz RJ, Sprang SR, Sambrook JF, Gething MJ. Affinity panning of a library of peptides displayed on bacteriophages reveals the binding specificity of BiP. Cell. 1993;75:717–728. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous