Identification of a lytic-cycle Epstein-Barr virus gene product that can regulate PKR activation - PubMed (original) (raw)
Identification of a lytic-cycle Epstein-Barr virus gene product that can regulate PKR activation
Jeremy Poppers et al. J Virol. 2003 Jan.
Abstract
The Epstein-Barr virus (EBV) SM protein is a posttranscriptional regulator of viral gene expression. Like many transactivators encoded by herpesviruses, SM transports predominantly unspliced viral mRNA cargo from the nucleus to the cytosol, where it is subsequently translated. This activity likely involves a region of the protein that has homology to the herpes simplex virus type 1 (HSV-1) ICP27 gene product, the first member of this class of regulators to be discovered. However, SM also contains a repetitive segment rich in arginine and proline residues that is dispensable for its effects on RNA transport and splicing. This portion of SM, comprised of RXP triplet repeats, shows homology to the carboxyl-terminal domain of Us11, a double-stranded RNA (dsRNA) binding protein encoded by HSV-1 that inhibits activation of the cellular PKR kinase. To evaluate the intrinsic ability of SM to regulate PKR, we expressed and purified several SM protein derivatives and examined their activity in a variety of biochemical assays. The full-length SM protein bound dsRNA, associated physically with PKR, and prevented PKR activation. Removal of the 37-residue RXP domain significantly compromised all of these activities. Furthermore, the SM RXP domain was itself sufficient to inhibit PKR activation and interact with the kinase. Relative to its Us11 counterpart, the SM RXP segment bound dsRNA with reduced affinity and responded differently to single-stranded competitor polynucleotides. Thus, SM represents the first EBV gene product expressed during the lytic cycle that can prevent PKR activation. In addition, the RXP repeat segment appears to be a conserved herpesvirus motif capable of associating with dsRNA and modulating activation of the PKR kinase, a molecule important for the control of translation and the cellular antiviral response.
Figures
FIG. 1.
EBV SM protein and Us11 polypeptide encoded by alphaherpesviruses both contain a repetitive RXP segment. (A) Domain structure of the SM and Us11 proteins. The 479-amino-acid SM polypeptide is encoded by a spliced mRNA that joins a segment of the BSLF2 open reading frame (S; amino acids 1 to 20) to the BMLF1 open reading frame (M; amino acids 41 to 479). A dotted line between the S segment and the M open reading frame represents the 20 residues encoded by sequences immediately upstream of the BMLF1 ATG codon. The RXP segment, along with the nuclear export sequence (NES) and ICP27 homology domain, is shown. The HSV-1 Us11 protein (Patton strain) is 155 amino acids long and contains a 68-amino-acid RXP domain in its carboxyl terminus. (B) Sequences of the RXP segments from SM appear aligned with various alphaherpesvirus Us11 proteins (HSV-1 Patton strain, HSV-1 strain 17, HSV-2 strain HG52, and simian virus B).
FIG. 2.
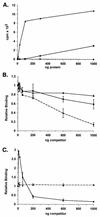
Binding of herpesvirus RXP domains to poly(IC). (A) Increasing quantities of purified GST or purified proteins containing either the HSV-1 Us11 RXP domain (GSTΔ1-87) or the EBV SM RXP segment (GST-SM-RXP) fused to GST were incubated with labeled poly(IC), and the amount of radioactivity retained after filtering the reaction mixtures through nitrocellulose was quantified. GSTΔ1-87, ▴; GST-SM-RXP, ▪; GST, •, broken line. (B) GST-SM-RXP (300 ng) was incubated with labeled poly(IC) (approximately 4 ng) in the presence of increasing amounts of excess unlabeled poly(IC) (•, broken line), poly(A) (▴), or poly(U) (▪). The amount of radioactivity retained on a nitrocellulose filter in the absence of unlabeled competitor was normalized to 1.0. (C) GSTΔ1-87 (10 ng) (▴, broken line) or GST-SM-RXP (300 ng) (•) was incubated with labeled poly(IC) in the presence of increasing amounts of unlabeled poly(C). Binding was quantified and normalized as described for panel B.
FIG. 3.
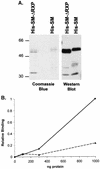
RXP segment embedded in full-length SM protein is important for dsRNA binding. (A) Full-length His-tagged SM (His-SM) and a mutant derivative that lacks the RXP domain (His-SM-ΔRXP) were isolated from cells infected with recombinant baculoviruses. Purified proteins were subjected to electrophoresis in SDS-polyacrylamide gels and either stained with Coomassie brilliant blue R-250 or transferred to a membrane which was subsequently probed with an anti-His monoclonal antibody. Positions of molecular size markers are shown (in kilodaltons). (B) A labeled 81-bp RNA duplex was prepared and incubated with increasing amounts of either His-SM (•) or His-S-ΔRXP (▴, broken line). Binding to dsRNA was measured by quantifying the radioactivity retained on the nitrocellulose filter.
FIG. 4.
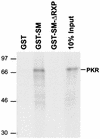
SM RXP domain is sufficient to interact with PKR. Purified GST fusion proteins bound to glutathione-agarose were incubated with in vitro-translated 35S-labeled PKR. Prior to mixing, both the lysate and the immobilized GST fusion proteins were treated separately with micrococcal nuclease. After the beads were collected and washed, the bound proteins were fractionated in SDS-polyacrylamide gels that were subsequently impregnated with a fluorophore and exposed to film. Positions of molecular size markers are indicated to the left (in kilodaltons). (A) GST-SM-RXP contains the 37-amino-acid segment from SM fused to GST; GSTΔ1-87 contains the carboxyl-terminal 68 amino acids from Us11 fused to GST; and GSTΔ88-155 contains the amino-terminal 87 amino acids of Us11 fused to GST. (B) The RXP domain does not interact nonspecifically with in vitro-translated luciferase or the glucocorticoid receptor. Full-length translation products are indicated with an asterisk to the right of each image.
FIG. 5.
Interaction of full-length SM with PKR requires the RXP repetitive segment. Purified GST fusion proteins were bound to glutathione-agarose and treated as described in the legend to Fig. 4. GST-SM contains amino acids 1 to 479 of SM fused to GST; GST-SM-ΔRXP contains an internal deletion of 37 amino acids that removes the RXP segment.
FIG. 6.
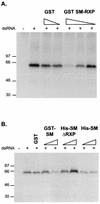
SM can regulate PKR activation. (A) The SM RXP segment is sufficient to inhibit PKR activation. Increasing amounts of either purified GST-RXP (9, 6, 32, or 85 pmol) or GST (37 or 98 pmol) were incubated in S10 extracts containing [γ-32P]ATP in the presence or absence of dsRNA. Following incubation at 30°C for 30 min, PKR was immunoprecipitated, and the immune complexes were fractionated in an SDS-polyacrylamide gel. The fixed, dried gel was subsequently exposed to film. Positions of molecular size markers are indicated to the left (in kilodaltons). (B) Removal of the RXP segment from SM reduces its ability to inhibit PKR activation. Same as in panel A except that the reaction mixtures contained purified GST-SM (3.75 or 12.5 pmol), His-SM (5.7 or 19 pmol), His-SM-ΔRXP (5.7 or 19 pmol), or GST (37 pmol). Positions of molecular size markers are indicated to the left (in kilodaltons).
Similar articles
- Regulation of PACT-Mediated Protein Kinase Activation by the OV20.0 Protein of Orf Virus.
Tseng YY, Liao GR, Sen GC, Lin FY, Hsu WL. Tseng YY, et al. J Virol. 2015 Nov;89(22):11619-29. doi: 10.1128/JVI.01739-15. Epub 2015 Sep 9. J Virol. 2015. PMID: 26355092 Free PMC article. - Activation of PKR: an open and shut case?
Cole JL. Cole JL. Trends Biochem Sci. 2007 Feb;32(2):57-62. doi: 10.1016/j.tibs.2006.12.003. Epub 2006 Dec 29. Trends Biochem Sci. 2007. PMID: 17196820 Free PMC article. Review. - Viral proteins targeting host protein kinase R to evade an innate immune response: a mini review.
Dzananovic E, McKenna SA, Patel TR. Dzananovic E, et al. Biotechnol Genet Eng Rev. 2018 Apr;34(1):33-59. doi: 10.1080/02648725.2018.1467151. Epub 2018 May 2. Biotechnol Genet Eng Rev. 2018. PMID: 29716441 Review.
Cited by
- RNA recognition by PKR during DNA virus infection.
Zhang R, Karijolich J. Zhang R, et al. J Med Virol. 2024 Feb;96(2):e29424. doi: 10.1002/jmv.29424. J Med Virol. 2024. PMID: 38285432 Review. - The Role of RNA Sensors in Regulating Innate Immunity to Gammaherpesviral Infections.
Zhang H, Sandhu PK, Damania B. Zhang H, et al. Cells. 2023 Jun 17;12(12):1650. doi: 10.3390/cells12121650. Cells. 2023. PMID: 37371120 Free PMC article. Review. - Mechanism of herpesvirus protein kinase UL13 in immune escape and viral replication.
Zhou L, Cheng A, Wang M, Wu Y, Yang Q, Tian B, Ou X, Sun D, Zhang S, Mao S, Zhao XX, Huang J, Gao Q, Zhu D, Jia R, Liu M, Chen S. Zhou L, et al. Front Immunol. 2022 Nov 30;13:1088690. doi: 10.3389/fimmu.2022.1088690. eCollection 2022. Front Immunol. 2022. PMID: 36531988 Free PMC article. Review. - Inhibition of PKR by Viruses.
Cesaro T, Michiels T. Cesaro T, et al. Front Microbiol. 2021 Oct 25;12:757238. doi: 10.3389/fmicb.2021.757238. eCollection 2021. Front Microbiol. 2021. PMID: 34759908 Free PMC article. Review. - The herpesvirus accessory protein γ134.5 facilitates viral replication by disabling mitochondrial translocation of RIG-I.
Liu X, Ma Y, Voss K, van Gent M, Chan YK, Gack MU, Gale M Jr, He B. Liu X, et al. PLoS Pathog. 2021 Mar 26;17(3):e1009446. doi: 10.1371/journal.ppat.1009446. eCollection 2021 Mar. PLoS Pathog. 2021. PMID: 33770145 Free PMC article.
References
- Bello, L. J., A. J. Davison, M. A. Glenn, A. Whitehouse, N. Rethmeier, T. F. Schulz, and J. Barklie Clements. 1999. The human herpesvirus-8 open reading frame 57 gene and its properties. J. Gen. Virol. 80:3207-3215. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources