Mechanism of cargo selection in the cytoplasm to vacuole targeting pathway - PubMed (original) (raw)
Mechanism of cargo selection in the cytoplasm to vacuole targeting pathway
Takahiro Shintani et al. Dev Cell. 2002 Dec.
Abstract
The proper functioning of eukaryotic organelles is largely dependent on the specific packaging of cargo proteins within transient delivery vesicles. The cytoplasm to vacuole targeting (Cvt) pathway is an autophagy-related trafficking pathway whose cargo proteins, aminopeptidase I and alpha-mannosidase, are selectively transported from the cytoplasm to the lysosome-like vacuole in yeast. This study elucidates a molecular mechanism for cargo specificity in this pathway involving four discrete steps. The Cvt19 receptor plays a central role in this process: distinct domains in Cvt19 recognize oligomerized cargo proteins and link them to the vesicle formation machinery via interaction with Cvt9 and Aut7. Because autophagy is the primary mechanism for organellar turnover, these results offer insights into physiological processes that are critical in cellular homeostasis, including specific packaging of damaged or superfluous organelles for lysosomal delivery and breakdown.
Figures
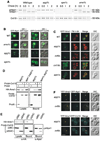
Figure 1. GFP-Ape1 Travels to the Vacuole via the Cvt Pathway
(A) Cell extracts from ape1 A (lanes 1–3), _pep4_Δ _ape1_Δ (lanes 4–6), _apg1_Δ _ape1_Δ (lane 7), and _cvt19_Δ _ape1_Δ (lane 8) strains containing either empty vector (pRS414; lanes 1 and 4), pApe1 (lanes 2 and 5), or pGFP-Ape1 (lanes 3, 6, 7, and 8) were resolved by SDS-PAGE. Western blots were performed with anti-Ape1 antiserum. (B) Western blot of cell extracts from _pep4_Δ _ape1_Δ (lane 1), _pep4_Δ (lane 2), _ape1_Δ (lane 3), and wild-type (lane 4) strains containing pGFP-Ape1 with anti-Ape1 (left panel) and anti-GFP (right panel) antisera. An asterisk (*) indicates nonspecific bands. (C) Localization of Ape1 by fluorescence microscopy of wild-type, _pep4_Δ, _apg1_Δ, and _cvt19_Δ strains expressing GFP-Ape1. Cells were grown to midlog phase and labeled with FM 4-64 in SCD medium. Bar, 5 µm. (D) CFP-Ape1 colocalizes with YFP-Aut7 in _apg1_Δ cells, but not in _cvt19_Δ cells. The _apg1_Δ and _cvt19_Δ cells coexpressing CFP-Ape1 and YFP-Aut7 were grown in SCD medium to midlog phase and examined with fluorescence and DIC microscopy. Bar, 5 µm.
Figure 2. Precursor Ape1 Propeptide Is Required for the Formation of the Ape1 Complex and Interaction With Cvt19
(A) Localization of the propeptide mutants of GFP-Ape1 by fluorescence and DIC microscopy. The _ape1_Δ cells harboring pGFP-Ape1, pGFP-Ape1Δ9–11, or pGFP-Ape1P22L were grown to midlog phase and labeled with FM 4-64 in SCD medium. Bar, 5 µm. (B) Two-hybrid analysis of the physical interaction between Ape1 mutants and Cvt19. Wild-type (PJ69-4A) and isogenic _ape1_Δ (YTS110) test strains were transformed with the activation domain (AD) plasmid containing CVT19 and the binding domain (BD) plasmid containing wild-type or mutant APE1. Transformants were grown on an SC-Leu-Ura plate and replica plated on an SC-Ade-Leu-Ura plate to assess the interaction-dependent activation of the ADE2 gene. The cells shown here were grown at 30°C for 2 days. (C) Interaction between prApe1 and Cvt19 is mediated by the prApe1 propeptide. Total lysate from the _ape1_Δ _apg1_Δ cells expressing HA-Cvt19 and either wild-type or mutant prApe1 from single-copy plasmids under the control of their own promoters were used for immunoprecipitation with anti-HA antibody, and the immunoprecipitates were analyzed by Western blot with anti-Ape1 and anti-HA antisera (IP). The top panel (Lysate) shows the protein blot of total cell lysate probed with anti-Ape1 antiserum.
Figure 3. Mapping Functional Domains within Cvt19
(A) Mapping the prApe1 binding domain by a yeast two-hybrid system. The _cvt19_Δ test strain (YTS111) was transformed with the binding domain plasmid containing the entire APE1 ORF and the activation domain plasmid containing mutant CVT19. The interaction was assessed by measuring the interaction-dependent induction of β-galactosidase activity. CC, coiled coil; ND, not determined. (B) Total lysates from the _apg1_Δ _cvt19_Δ cells expressing either wild-type or mutant Cvt19 fused with protein A (ProtA-Cvt19) were used to precipitate ProtA-Cvt19 proteins with IgG Sepharose. The left and right panels show the protein blots of total cell extracts (Lysate) and the IgG precipitates (Bound), respectively, which were probed with anti-Ape1 antiserum. Single arrowheads show ProtA-Cvt19 derivatives, and a double arrowhead indicates protein A without fusion. Although Cvt19 mutants containing the coiled-coil domain (Δ10C, Δ15C, Δ20C, Δ28C, Δ21–28C, Δ224C, and Δ152N) were all able to precipitate prApe1, only the results from Δ224C and Δ152N mutants were shown in this figure. (C) Protein blots of total lysates from the _cvt19_Δ cells expressing Cvt19 deletion mutants fused with GFP probed with anti-Ape1 (upper panel) and anti-GFP (lower panel) antisera. An asterisk (*) indicates nonspecific bands. (D) Localization of wild-type and mutant Cvt19 proteins by fluorescence and DIC microscopy of _cvt19_Δ strains expressing wild-type or mutant GFP-Cvt19. Cells were grown to midlog phase and labeled with FM 4-64 in SCD medium. Bar, 5 µm. (E) Colocalization of YFP-Cvt19 and YFP-Cvt19Δ20C with CFP-Ape1 and CFP-Aut7 in a _cvt19_Δ strain by fluorescence and DIC microscopy. Bar, 5 µm.
Figure 4. Cvt9 Facilitates the Localization of the Ape1-Cvt19 Complex to the Preautophagosomal Structure
(A) Mapping of Aut7 and Cvt9 binding domains within Cvt19 by a yeast two-hybrid system. The _cvt19_Δ test strain (WHY4) was transformed with the binding domain plasmid containing the entire CVT9 or AUT7 ORF (pGBD-Cvt9 or pGBD-Aut7, respectively) and the activation domain plasmid containing wild-type or mutant CVT19. Transformants were selected on SC-Leu-Ura plates (data not shown) and replicated to SC-Ade plates. The figures show cells grown at 30°C for 2 days. (B) A protein A-Aut7ΔR affinity isolation experiment was performed in lysates from the _apg1_Δ _cvt19_Δ strain expressing full-length Cvt19 or the indicated truncate. C-terminal deletion of Cvt19 compromised its coisolation with Aut7. (C) Cvt9 interacts with the C terminus of Cvt19. Total lysates from _apg1_Δ _cvt19_Δ cells expressing HA-Cvt9 and either wild-type or mutant Cvt19 fused with protein A (ProtA-Cvt19) were used to precipitate ProtA-Cvt19 proteins with IgG Sepharose. The left and right panels show the protein blots of total cell lysates (Lysate) and the IgG precipitates (Bound), respectively, which were probed with anti-HA antibody. (D and E) Localization of GFP-Ape1 (D) or GFP-Cvt19 (E) in the _aut7_Δ, _cvt9_Δ, and _aut7_Δ _cvt9_Δ strains by fluorescence and DIC microscopy. Bar, 5 µm. (F and G) Colocalization of CFP-Ape1 with YFP-Aut7 (F) or YFP-Cvt19 (G) in the _cvt9_Δ strain by fluorescence and DIC microscopy. Bar, 5 µm.
Figure 5. Precursor Ape1 Is Required for the Efficient Transport of Cvt19 and the Cargo Protein Ams1
(A) Wild-type, _apg1_Δ, _ape1_Δ, and _ams1_Δ cells were pulse labeled for 10 min in SMD medium and chased for the indicated times in SMD medium. Ape1 and Cvt19 were immunoprecipitated from cell extracts and analyzed by SDS-PAGE. (B) Localization of GFP-Ape1 or GFP-Cvt19 in wild-type, _ams1_Δ, _apg1_Δ, and _ape1_Δ strains by fluorescence and DIC microscopy. Bar, 5 µm. (C) Localization of GFP-Ams1 in wild-type, _apg1_Δ, _cvt9_Δ, _cvt19_Δ, and _ape1_Δ strains by fluorescence and DIC microscopy. Cells were grown to midlog phase and labeled with FM 4-64 in SCD medium. Bar, 5 µm. (D) Ams1 associates with Cvt19, independent of Ape1. The _apg1_Δ or _ape1_Δ _apg1_Δ cells were transformed with pHA-Ams1 and pRS416-CuProtA or pCuProtA-Cvt19. IgG Sepharose was used to precipitate ProtA-Cvt19 from cell lysates. The left and right panels show the protein blots of total cell lysates (Lysate) and the IgG precipitates (Bound), respectively, which were probed with anti-HA antibody. (E) The Ams1 binding site in Cvt19 is different from the prApe1 binding site. Total lysates from _apg1_Δ _cvt19_Δ cells expressing HA-Ams1 and either ProtA-Cvt19Δ28C or ProtA-Cvt19Δ224C were used to precipitate ProtA-Cvt19 proteins with IgG Sepharose. Total cell lysates (Lysate) and the IgG precipitates (Bound) were analyzed by Western blot with anti-HA or anti-Ape1 antisera. (F) Colocalization of YFP-Ams1 with CFP-Ape1 or CFP-Cvt19 in _apg1_Δ and _cvt9_Δ cells by fluorescence and DIC microscopy. Bar, 5 µm.
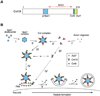
Figure 6. Model for Selective Targeting of Cvt Cargos into the Cvt Vesicle
(A) Schematic drawing of binding domains in Cvt19. The prApe1 binding domain (blue) is located in the region from amino acid residues 153 to 191, which contains the predicted coiled-coil motif. The 10-amino acid region from 406 to 415 (yellow) and the 8-amino acid region from 388 to 395 (green) are for Aut7 binding and Cvt9 binding, respectively. An exact binding domain for Ams1 has not been determined but is located in a region within amino acid residues 192-387 (red arrow). (B) After prApe1 is synthesized it immediately forms dodecamers in the cytosol, which are further assembled into an Ape1 complex dependent on its propeptide. Cvt19 then binds to the Ape1 complex, which is also mediated by the prApe1 propeptide. Because Ams1 is able to associate with Cvt19 via a domain distinct from the prApe1 binding domain, Ams1 could be concentrated on the Ape1 complex as a consequence of the Ape1-Cvt19 complex formation. Cvt9 then interacts with the C terminus of Cvt19 to recruit the Ape1-Cvt19-Ams1 complex to the PAS, where Aut7 binds to the extreme C terminus of Cvt19 to ensure the incorporation of the complex into the Cvt vesicle. See text for details.
Similar articles
- Studies of cargo delivery to the vacuole mediated by autophagosomes in Saccharomyces cerevisiae.
Suzuki K, Kamada Y, Ohsumi Y. Suzuki K, et al. Dev Cell. 2002 Dec;3(6):815-24. doi: 10.1016/s1534-5807(02)00359-3. Dev Cell. 2002. PMID: 12479807 - Cargo proteins facilitate the formation of transport vesicles in the cytoplasm to vacuole targeting pathway.
Shintani T, Klionsky DJ. Shintani T, et al. J Biol Chem. 2004 Jul 16;279(29):29889-94. doi: 10.1074/jbc.M404399200. Epub 2004 May 11. J Biol Chem. 2004. PMID: 15138258 Free PMC article. - Atg11 links cargo to the vesicle-forming machinery in the cytoplasm to vacuole targeting pathway.
Yorimitsu T, Klionsky DJ. Yorimitsu T, et al. Mol Biol Cell. 2005 Apr;16(4):1593-605. doi: 10.1091/mbc.e04-11-1035. Epub 2005 Jan 19. Mol Biol Cell. 2005. PMID: 15659643 Free PMC article. - Autophagy in yeast: a review of the molecular machinery.
Huang WP, Klionsky DJ. Huang WP, et al. Cell Struct Funct. 2002 Dec;27(6):409-20. doi: 10.1247/csf.27.409. Cell Struct Funct. 2002. PMID: 12576634 Review. - Transport of proteins to the yeast vacuole: autophagy, cytoplasm-to-vacuole targeting, and role of the vacuole in degradation.
Teter SA, Klionsky DJ. Teter SA, et al. Semin Cell Dev Biol. 2000 Jun;11(3):173-9. doi: 10.1006/scdb.2000.0163. Semin Cell Dev Biol. 2000. PMID: 10906274 Review.
Cited by
- Peroxisomal Pex3 activates selective autophagy of peroxisomes via interaction with the pexophagy receptor Atg30.
Burnett SF, Farré JC, Nazarko TY, Subramani S. Burnett SF, et al. J Biol Chem. 2015 Mar 27;290(13):8623-31. doi: 10.1074/jbc.M114.619338. Epub 2015 Feb 18. J Biol Chem. 2015. PMID: 25694426 Free PMC article. - Recruitment of Atg9 to the preautophagosomal structure by Atg11 is essential for selective autophagy in budding yeast.
He C, Song H, Yorimitsu T, Monastyrska I, Yen WL, Legakis JE, Klionsky DJ. He C, et al. J Cell Biol. 2006 Dec 18;175(6):925-35. doi: 10.1083/jcb.200606084. J Cell Biol. 2006. PMID: 17178909 Free PMC article. - Vps51 is part of the yeast Vps fifty-three tethering complex essential for retrograde traffic from the early endosome and Cvt vesicle completion.
Reggiori F, Wang CW, Stromhaug PE, Shintani T, Klionsky DJ. Reggiori F, et al. J Biol Chem. 2003 Feb 14;278(7):5009-20. doi: 10.1074/jbc.M210436200. Epub 2002 Nov 20. J Biol Chem. 2003. PMID: 12446664 Free PMC article. - A comprehensive glossary of autophagy-related molecules and processes (2nd edition).
Klionsky DJ, Baehrecke EH, Brumell JH, Chu CT, Codogno P, Cuervo AM, Debnath J, Deretic V, Elazar Z, Eskelinen EL, Finkbeiner S, Fueyo-Margareto J, Gewirtz D, Jäättelä M, Kroemer G, Levine B, Melia TJ, Mizushima N, Rubinsztein DC, Simonsen A, Thorburn A, Thumm M, Tooze SA. Klionsky DJ, et al. Autophagy. 2011 Nov;7(11):1273-94. doi: 10.4161/auto.7.11.17661. Epub 2011 Nov 1. Autophagy. 2011. PMID: 21997368 Free PMC article. - History of the Selective Autophagy Research: How Did It Begin and Where Does It Stand Today?
Kirkin V. Kirkin V. J Mol Biol. 2020 Jan 3;432(1):3-27. doi: 10.1016/j.jmb.2019.05.010. Epub 2019 May 11. J Mol Biol. 2020. PMID: 31082435 Free PMC article. Review.
References
- Adams A, Gottschling DE, Kaiser CA, Stearns T. Methods in Yeast Genetics. A Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1998.
- Brunk UT, Terman A. The mitochondrial-lysosomal axis theory of aging: accumulation of damaged mitochondria as a result of imperfect autophagocytosis. Eur. J. Biochem. 2002;269:1996–2002. - PubMed
- Gerhardt B, Kordas TJ, Thompson CM, Patel P, Vida T. The vesicle transport protein Vps33p is an ATP-binding protein that localizes to the cytosol in an energy-dependent manner. J. Biol. Chem. 1998;273:15818–15829. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM053396/GM/NIGMS NIH HHS/United States
- R01 GM053396-12/GM/NIGMS NIH HHS/United States
- R01 GM053396-13/GM/NIGMS NIH HHS/United States
- GM53396/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases