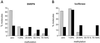The DNA methyltransferase-like protein DNMT3L stimulates de novo methylation by Dnmt3a - PubMed (original) (raw)
The DNA methyltransferase-like protein DNMT3L stimulates de novo methylation by Dnmt3a
Frederic Chedin et al. Proc Natl Acad Sci U S A. 2002.
Abstract
Dnmt3L is required for the establishment of maternal methylation imprints at imprinting centers (ICs). Dnmt3L, however, lacks the conserved catalytic domain common to DNA methyltransferases. In an attempt to define its function, we coexpressed DNMT3L with each of the two known de novo methyltransferases, Dnmt3a and DNMT3B, in human cells and monitored de novo methylation by using replicating minichromosomes carrying various ICs as targets. Coexpression of DNMT3L with DNMT3B led to little or no change in target methylation. However, coexpression of DNMT3L with Dnmt3a resulted in a striking stimulation of de novo methylation by Dnmt3a. Stimulation was observed at maternally methylated ICs such as small nuclear ribonucleoprotein polypeptide N (SNRPN), Snrpn, and Igf2rAir, as well as at various nonimprinted sequences present on the episomes. Stimulation of Dnmt3a by DNMT3L was also observed at endogenous sequences in the genome. Therefore, DNMT3L acts as a general stimulatory factor for de novo methylation by Dnmt3a. The implications of these findings for the function of DNMT3L and Dnmt3a in DNA methylation and genomic imprinting are discussed.
Figures
Fig 1.
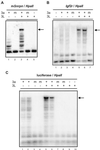
DNMT3L stimulates de novo methylation by Dnmt3a but not by DNMT3B at the SNRPN IC. DNA harvested from human 293/EBNA1 cells carrying pFC19 or pFC30 (in which a key ≈1-kb region of the SNRPN IC was cloned in either orientation) was digested with an excess of the methylation-sensitive enzyme _Hpa_II. The resulting DNA species were separated by electrophoresis through an agarose gel, transferred to a nylon membrane, and hybridized with a probe corresponding to the SNRPN IC. Two bands (684 and 754 bp for pFC19, and 684 and 221 bp for pFC30) were expected if no methylation was present. CpG methylation prevents cleavage by _Hpa_II and results in higher molecular weight species that can be easily separated. (A) Each target plasmid was introduced into cells alone, together with an expression vector for Dnmt3a, or together with expression vectors for both Dnmt3a and DNMT3L, as indicated. (B) Plasmid pFC19 was introduced in cells either with DNMT3B or with both DNMT3B and DNMT3L expression vectors, as indicated.
Fig 2.
DNMT3L stimulates de novo methylation by Dnmt3a at maternally methylated ICs and nonimprinted sequences. Key regions of the murine Snrpn IC (A) and murine Igf2r/Air IC (B) carried by pFC49 and pFC60, respectively, were introduced into cells together with expression vectors for Dnmt3a and/or DNMT3L, as indicated. (C) pCLH22, which carries the luciferase gene, was used. The probes used to detect methylation correspond to the respective ICs or to the luciferase gene.
Fig 3.
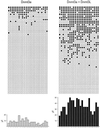
Coexpression of Dnmt3a and DNMT3L results in stimulation of methylation at all CpG sites. Methylation at the SNRPN locus was analyzed by using bisulfite genomic sequencing on samples recovered from cells carrying pFC19 and expressing Dnmt3a alone (Left) or Dnmt3a and DNMT3L together (Right). The distribution of methylated sites (•) at each of the 23 CpG sites along the top strand of the SNRPN IC is depicted. A histogram representing the total number of methylation events observed at a given site for the whole sample is shown at the bottom. Sample size was identical for both categories (53 independent molecules analyzed per group).
Fig 4.
Coexpression of Dnmt3a and DNMT3L results in extensive DNA methylation patterns. The percentage of CpG methylation observed at the SNRPN IC (A) or at the luciferase gene (B) was determined in individual molecules analyzed by bisulfite sequencing. Molecules were grouped into five classes depending on the percentage of methylation observed, as shown on the x axis. The fraction of each class within the total sample size is indicated on the y axis. Methylation of the target region resulted from Dnmt3a alone (gray bars) or Dnmt3a and DNMT3L together (black bars).
Fig 5.
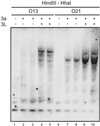
Stimulation by DNMT3L is detected at the luciferase gene integrated in the genome. Two cell lines (O13 and O21) carrying the luciferase gene at distinct loci were transfected with Dnmt3a or Dnmt3a + DNMT3L expression vectors. After transfection (3.5 days), genomic DNA was harvested and digested with excess _Hin_dIII and _Hha_I, a methylation-sensitive enzyme. The resulting DNA species were then separated by agarose gel electrophoresis, Southern transferred onto a membrane, and hybridized to a probe complementary to the luciferase gene. The presence of Dnmt3a and/or DNMT3L expression vectors was as indicated. CpG methylation prevents cleavage by _Hha_I and results in higher molecular weight species that can be easily distinguished.
Similar articles
- Physical and functional interactions between the human DNMT3L protein and members of the de novo methyltransferase family.
Chen ZX, Mann JR, Hsieh CL, Riggs AD, Chédin F. Chen ZX, et al. J Cell Biochem. 2005 Aug 1;95(5):902-17. doi: 10.1002/jcb.20447. J Cell Biochem. 2005. PMID: 15861382 - Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation.
Jia D, Jurkowska RZ, Zhang X, Jeltsch A, Cheng X. Jia D, et al. Nature. 2007 Sep 13;449(7159):248-51. doi: 10.1038/nature06146. Epub 2007 Aug 22. Nature. 2007. PMID: 17713477 Free PMC article. - DNMT3L stimulates the DNA methylation activity of Dnmt3a and Dnmt3b through a direct interaction.
Suetake I, Shinozaki F, Miyagawa J, Takeshima H, Tajima S. Suetake I, et al. J Biol Chem. 2004 Jun 25;279(26):27816-23. doi: 10.1074/jbc.M400181200. Epub 2004 Apr 21. J Biol Chem. 2004. PMID: 15105426 - Domain Structure of the Dnmt1, Dnmt3a, and Dnmt3b DNA Methyltransferases.
Tajima S, Suetake I, Takeshita K, Nakagawa A, Kimura H. Tajima S, et al. Adv Exp Med Biol. 2016;945:63-86. doi: 10.1007/978-3-319-43624-1_4. Adv Exp Med Biol. 2016. PMID: 27826835 Review. - Dynamic expression of DNA methyltransferases (DNMTs) in oocytes and early embryos.
Uysal F, Akkoyunlu G, Ozturk S. Uysal F, et al. Biochimie. 2015 Sep;116:103-13. doi: 10.1016/j.biochi.2015.06.019. Epub 2015 Jul 2. Biochimie. 2015. PMID: 26143007 Review.
Cited by
- The N-terminal region of DNMT3A engages the nucleosome surface to aid chromatin recruitment.
Wapenaar H, Clifford G, Rolls W, Pasquier M, Burdett H, Zhang Y, Deák G, Zou J, Spanos C, Taylor MRD, Mills J, Watson JA, Kumar D, Clark R, Das A, Valsakumar D, Bramham J, Voigt P, Sproul D, Wilson MD. Wapenaar H, et al. EMBO Rep. 2024 Dec;25(12):5743-5779. doi: 10.1038/s44319-024-00306-3. Epub 2024 Nov 11. EMBO Rep. 2024. PMID: 39528729 Free PMC article. - Epigenetic DNA Methylation and Protein Homocysteinylation: Key Players in Hypertensive Renovascular Damage.
Ren L, Pushpakumar S, Almarshood H, Das SK, Sen U. Ren L, et al. Int J Mol Sci. 2024 Oct 29;25(21):11599. doi: 10.3390/ijms252111599. Int J Mol Sci. 2024. PMID: 39519150 Free PMC article. Review. - Using human disease mutations to understand de novo DNA methyltransferase function.
Rolls W, Wilson MD, Sproul D. Rolls W, et al. Biochem Soc Trans. 2024 Oct 30;52(5):2059-2075. doi: 10.1042/BST20231017. Biochem Soc Trans. 2024. PMID: 39446312 Free PMC article. Review. - Imprinting as Basis for Complex Evolutionary Novelties in Eutherians.
Schuff M, Strong AD, Welborn LK, Ziermann-Canabarro JM. Schuff M, et al. Biology (Basel). 2024 Aug 31;13(9):682. doi: 10.3390/biology13090682. Biology (Basel). 2024. PMID: 39336109 Free PMC article. Review. - Cancer-associated DNA hypermethylation of Polycomb targets requires DNMT3A dual recognition of histone H2AK119 ubiquitination and the nucleosome acidic patch.
Gretarsson KH, Abini-Agbomson S, Gloor SL, Weinberg DN, McCuiston JL, Kumary VUS, Hickman AR, Sahu V, Lee R, Xu X, Lipieta N, Flashner S, Adeleke OA, Popova IK, Taylor HF, Noll K, Windham CL, Maryanski DN, Venters BJ, Nakagawa H, Keogh MC, Armache KJ, Lu C. Gretarsson KH, et al. Sci Adv. 2024 Aug 30;10(35):eadp0975. doi: 10.1126/sciadv.adp0975. Epub 2024 Aug 28. Sci Adv. 2024. PMID: 39196936 Free PMC article.
References
- Bestor T. H. (2000) Hum. Mol. Genet. 9, 2395-2402. - PubMed
- Okano M., Bell, D. W., Haber, D. A. & Li, E. (1999) Cell 99, 247-257. - PubMed
- Bourc'his D., Xu, G. L., Lin, C. S., Bollman, B. & Bestor, T. H. (2001) Science 294, 2536-2539. - PubMed
- Hata K., Okano, M., Lei, H. & Li, E. (2002) Development (Cambridge, U.K.) 129, 1983-1993. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases