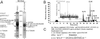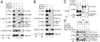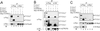CUL7: A DOC domain-containing cullin selectively binds Skp1.Fbx29 to form an SCF-like complex - PubMed (original) (raw)
CUL7: A DOC domain-containing cullin selectively binds Skp1.Fbx29 to form an SCF-like complex
Dora C Dias et al. Proc Natl Acad Sci U S A. 2002.
Abstract
Selective protein degradation targeted by members of the F-box protein family plays pivotal roles in cell biology. It is widely accepted that an F-box protein directs substrate ubiquitination within a Skp1.CUL1.F-box protein.ROC1 (SCF-ROC1) E3 ubiquitin ligase complex. This assembly utilizes the CUL1 molecular scaffold, allowing the F-box protein to position its bound substrate for ubiquitination by a ROC1-recruited E2-conjugating enzyme. Here, we describe an alternative mechanism for assembling an F-box protein-based E3 complex through a previously uncharacterized cullin, CUL7, identified by mass spectrometry as a ROC1-interacting protein. CUL7 is a large polypeptide containing a cullin domain, which is responsible for ROC1 binding, and a DOC domain, which is also present in the anaphase-promoting complex. Remarkably, CUL7 assembles an SCF-ROC1-like E3 ubiquitin ligase complex consisting of Skp1, CUL7, the Fbx29 F-box protein, and ROC1. In contrast to CUL1 that binds Skp1 by itself, CUL7 interacts with the Skp1.Fbx29 complex, but not with Skp1 alone. Strikingly, CUL7 selectively interacts with Skp1.Fbx29 but not with Skp1.betaTRCP2 or Skp1.Skp2. Thus, CUL7 may define a previously uncharacterized, Fbx29-mediated, and ubiquitin-dependent proteolysis pathway.
Figures
Fig 1.
Identification of CUL7 as a ROC1-interacting protein. (A) Coomassie stain analysis of the Flag-ROC1 immunoprecipitates. Flag-peptide eluants derived from ≈20 150-mm plates' worth of cells expressing either Flag (F)-ROC1 (lane 1) or Flag (F)-ΔN-CUL4A (lane 2) were concentrated by TCA precipitation followed by electrophoresis through SDS/10% PAGE and Coomassie staining. (B and C) Identification of p200 as CUL7 (KIAA0076) by micro-HPLC/electrospray ionization ion trap (ESI-IT)-MS/MS analysis. p200-derived tryptic peptides were fractionated by micro-C18–HPLC; the elution profile is shown in B. The insertion in B is an MS spectrum showing the detection of a peptide ion with m/z 874.4. Database searches using the entire p200 MS/MS data resulted in the identification of KIAA0076 gene product, shown in C. A detailed description of the MS/MS analysis can be found in the supporting information.
Fig 2.
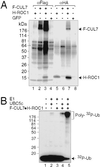
Structural domains within CUL7. Structural similarity between CUL7 and KIAA0708 is indicated. HERC2-HD refers to a CUL7 N-terminal region that spans 72 amino acids, is enriched with glycine and acidic residues, and shares 59% similarity to the corresponding sequences in HERC2. IBR represents an in-between RING finger domain. Segments A–F are representative mouse EST clones [GenBank database accession nos. (A), (B), (C), (D), (E), and (F)] that are highly homologous to CUL7.
Fig 3.
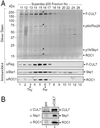
CUL7 forms a complex with ROC1 that supports Ub polymerization. (A) CUL7 interacts with ROC1. Cells (293T) were transfected with pCR3.1-Flag (F)-CUL7 and/or pcDNA-HA (H)-ROC1 as indicated. 35S-labeled extracts (≈3 mg of protein) were immunoprecipitated with αFlag (lanes 1–4) or αHA (lanes 5–8) antibodies, and the resulting immunoprecipitates were separated by SDS/4–20% PAGE followed by autoradiography. The identities of the polypeptides of molecular masses ranging from 50 to 80 kDa present in the anti-Flag immunoprecipitates (lanes 2 and 3) are presently unknown. (B) The CUL7⋅ROC1 complex supports ubiquitin polymerization. The Flag (F)-CUL7⋅HA (H)-ROC1 complex, immobilized onto the M2 beads, was analyzed for ubiquitin ligase activity as described (4). The autoradiogram is shown.
Fig 4.
Identification of the CUL7⋅Skp1⋅Fbx29⋅ROC1 complex. (A) Comigration of CUL7, ROC1, p19, and p64 upon Superdex-200 gel filtration. The Flag-peptide eluant containing Flag (F)-CUL7 and its associated proteins was separated by gel filtration on a Superdex-200 column, as described in the supporting information. Aliquots of the indicated fractions (12 μl) were electrophoresed on 4–20% gradient denaturing gels and analyzed by silver staining (Upper) and by immunoblotting by using antibodies as indicated (Lower). Arrows at the bottom denote the positions where size markers thyroglobulin (Thy, 669 kDa) and ferritin (Fer, 440 kDa) migrated. (B) Association of endogenous CUL7 with Skp1 and ROC1. Preimmune or anti-CUL7 serum, 100 μl each, was prebound to protein A agarose beads (15 μl) overnight at 4°C, followed by an extensive wash. The resulting beads then were mixed with extracts of 293 cells (≈72 mg of protein), and the mixture was rocked overnight at 4°C. After washing, the immunopurified proteins were eluted from the beads and analyzed by immunoblotting using antibodies as indicated.
Fig 5.
CUL7 binds Skp1⋅Fbx29 but not Skp1 alone. Cells (293T) were cotransfected with an indicated set of vectors expressing Flag (F)-CUL7, Flag (F)-CUL1, Skp1-HA (H), His-Skp1, Myc (M)-Fbx29, Myc (M)-Fbx29 ΔF, Flag (F)-ROC1, or HA (H)-ROC1. Protein extracts (10 mg) were immunoprecipitated by anti-Myc, anti-Flag, or anti-HA antibodies, as indicated. The resulting immunoprecipitates were analyzed by immunoblots by using the specified antibodies.
Fig 6.
Selective interaction between CUL7 and Skp1⋅Fbx29. For comparison, pCR3.1-Flag (F)-CUL7 (lanes 1 and 4) or pCR3.1-Flag (F)-CUL1 (lanes 2 and 5) were cotransfected with pCR3.1-Myc (M)-Fbx29 (A), pcDNA-HA (H)-βTRCP2 (B), or pcDNA-Myc (M)-Skp2 (C), along with pcDNA-Skp1 and pcDNA-HA (H)-ROC1 or pcDNA-Myc (M)-ROC1. Immunoprecipitates were analyzed by Western immunoblotting by using antibodies as specified. (B) The anti-Flag blot is shown with both long (Middle, 2-min) and short (Top, 15-s) exposures for better illustration of the inability of CUL7 to interact with Skp1⋅HA-βTRCP2. (C) Note that the Myc-Skp2 protein that was coimmunoprecipitated with Flag-CUL1 migrated slightly faster than the nonspecific IgG heavy chains (compare lane 2 with lanes 1 and 3, Middle). In addition, anti-Skp2 immunoblot of the Myc immunoprecipitates is not shown. For unknown reasons, anti-Skp2 antibodies used in this study exhibited unusually high levels of nonspecific interactions with the heavy chains of Myc antibodies, which comigrated with the Myc-Skp2 proteins, thereby precluding the evaluation of Myc-Skp2 in the immunoprecipitates.
Similar articles
- The SCF(HOS/beta-TRCP)-ROC1 E3 ubiquitin ligase utilizes two distinct domains within CUL1 for substrate targeting and ubiquitin ligation.
Wu K, Fuchs SY, Chen A, Tan P, Gomez C, Ronai Z, Pan ZQ. Wu K, et al. Mol Cell Biol. 2000 Feb;20(4):1382-93. doi: 10.1128/MCB.20.4.1382-1393.2000. Mol Cell Biol. 2000. PMID: 10648623 Free PMC article. - Phosphorylation- and Skp1-independent in vitro ubiquitination of E2F1 by multiple ROC-cullin ligases.
Ohta T, Xiong Y. Ohta T, et al. Cancer Res. 2001 Feb 15;61(4):1347-53. Cancer Res. 2001. PMID: 11245432 - Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex.
Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, Chu C, Koepp DM, Elledge SJ, Pagano M, Conaway RC, Conaway JW, Harper JW, Pavletich NP. Zheng N, et al. Nature. 2002 Apr 18;416(6882):703-9. doi: 10.1038/416703a. Nature. 2002. PMID: 11961546 - Genetically engineered mouse models for functional studies of SKP1-CUL1-F-box-protein (SCF) E3 ubiquitin ligases.
Zhou W, Wei W, Sun Y. Zhou W, et al. Cell Res. 2013 May;23(5):599-619. doi: 10.1038/cr.2013.44. Epub 2013 Mar 26. Cell Res. 2013. PMID: 23528706 Free PMC article. Review. - The SCF ubiquitin ligase: an extended look.
Jackson PK, Eldridge AG. Jackson PK, et al. Mol Cell. 2002 May;9(5):923-5. doi: 10.1016/s1097-2765(02)00538-5. Mol Cell. 2002. PMID: 12049727 Review.
Cited by
- Cul4A targets p27 for degradation and regulates proliferation, cell cycle exit, and differentiation during erythropoiesis.
Li B, Jia N, Kapur R, Chun KT. Li B, et al. Blood. 2006 Jun 1;107(11):4291-9. doi: 10.1182/blood-2005-08-3349. Epub 2006 Feb 7. Blood. 2006. PMID: 16467204 Free PMC article. - Targeted disruption of p185/Cul7 gene results in abnormal vascular morphogenesis.
Arai T, Kasper JS, Skaar JR, Ali SH, Takahashi C, DeCaprio JA. Arai T, et al. Proc Natl Acad Sci U S A. 2003 Aug 19;100(17):9855-60. doi: 10.1073/pnas.1733908100. Epub 2003 Aug 6. Proc Natl Acad Sci U S A. 2003. PMID: 12904573 Free PMC article. - RING-finger E3 ligases regulatory network in PI3K/AKT-mediated glucose metabolism.
Wang W, Shi B, Cong R, Hao M, Peng Y, Yang H, Song J, Feng D, Zhang N, Li D. Wang W, et al. Cell Death Discov. 2022 Aug 24;8(1):372. doi: 10.1038/s41420-022-01162-7. Cell Death Discov. 2022. PMID: 36002460 Free PMC article. Review. - Identifying biological pathways that underlie primordial short stature using network analysis.
Hanson D, Stevens A, Murray PG, Black GC, Clayton PE. Hanson D, et al. J Mol Endocrinol. 2014 Jun;52(3):333-44. doi: 10.1530/JME-14-0029. Epub 2014 Apr 7. J Mol Endocrinol. 2014. PMID: 24711643 Free PMC article. - RINGs of good and evil: RING finger ubiquitin ligases at the crossroads of tumour suppression and oncogenesis.
Lipkowitz S, Weissman AM. Lipkowitz S, et al. Nat Rev Cancer. 2011 Aug 24;11(9):629-43. doi: 10.1038/nrc3120. Nat Rev Cancer. 2011. PMID: 21863050 Free PMC article. Review.
References
- Zachariae W., Shevchenko, A., Andrews, P. D., Ciosk, R., Galova, M., Stark, M. J., Mann, M. & Nasmyth, K. (1998) Science 279, 1216-1219. - PubMed
- Yu H., Peters, J.-M., King, R. W., Page, A. M., Hieter, P. & Kirschner, M. W. (1998) Science 279, 1219-1222. - PubMed
- Skowyra D., Koepp, D. M., Kamura, T., Conrad, M. N., Conaway, R. C., Conaway, J. W., Elledge, S. J. & Harper, J. W. (1999) Science 284, 662-665. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous