Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress - PubMed (original) (raw)
Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress
Joel A Kreps et al. Plant Physiol. 2002 Dec.
Abstract
To identify genes of potential importance to cold, salt, and drought tolerance, global expression profiling was performed on Arabidopsis plants subjected to stress treatments of 4 degrees C, 100 mM NaCl, or 200 mM mannitol, respectively. RNA samples were collected separately from leaves and roots after 3- and 27-h stress treatments. Profiling was conducted with a GeneChip microarray with probe sets for approximately 8,100 genes. Combined results from all three stresses identified 2,409 genes with a greater than 2-fold change over control. This suggests that about 30% of the transcriptome is sensitive to regulation by common stress conditions. The majority of changes were stimulus specific. At the 3-h time point, less than 5% (118 genes) of the changes were observed as shared by all three stress responses. By 27 h, the number of shared responses was reduced more than 10-fold (< 0.5%), consistent with a progression toward more stimulus-specific responses. Roots and leaves displayed very different changes. For example, less than 14% of the cold-specific changes were shared between root and leaves at both 3 and 27 h. The gene with the largest induction under all three stress treatments was At5g52310 (LTI/COR78), with induction levels in roots greater than 250-fold for cold, 40-fold for mannitol, and 57-fold for NaCl. A stress response was observed for 306 (68%) of the known circadian controlled genes, supporting the hypothesis that an important function of the circadian clock is to "anticipate" predictable stresses such as cold nights. Although these results identify hundreds of potentially important transcriptome changes, the biochemical functions of many stress-regulated genes remain unknown.
Figures
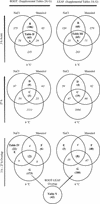
Figure 1
Venn diagrams showing an overview of stress-regulated changes (> 2-fold) for different treatment conditions. Stress-induced changes were identified in 2,678 probe sets (which approximately equals the number of genes). The number of changes and the percentage corresponding to up-regulation are shown. Numbers for fresh medium represent a comparison between 3 and 27 h. Numbers for the three stresses represent a comparison with the average of the 3- and 27-h control.
Figure 2
Venn diagrams showing the distribution of stimulus-specific and shared stress responses (> 2-fold). Numbers in italics correspond to probe sets identified in a specific category. Most of these changes were only observed once as a transient change. Non-italicized numbers in parentheses correspond to individual genes that passed a more stringent criterion for reproducibility (see text). Further information on these selected changes can be found in Tables II through V (shown here) or as supplemental information in Supplemental Tables 2A–G and 3A–G (They can be viewed at
).
Similar articles
- Transcriptomic identification of candidate genes involved in sunflower responses to chilling and salt stresses based on cDNA microarray analysis.
Fernandez P, Di Rienzo J, Fernandez L, Hopp HE, Paniego N, Heinz RA. Fernandez P, et al. BMC Plant Biol. 2008 Jan 26;8:11. doi: 10.1186/1471-2229-8-11. BMC Plant Biol. 2008. PMID: 18221554 Free PMC article. - Comprehensive transcriptional profiling of NaCl-stressed Arabidopsis roots reveals novel classes of responsive genes.
Jiang Y, Deyholos MK. Jiang Y, et al. BMC Plant Biol. 2006 Oct 12;6:25. doi: 10.1186/1471-2229-6-25. BMC Plant Biol. 2006. PMID: 17038189 Free PMC article. - Transcriptional profiling of Arabidopsis heat shock proteins and transcription factors reveals extensive overlap between heat and non-heat stress response pathways.
Swindell WR, Huebner M, Weber AP. Swindell WR, et al. BMC Genomics. 2007 May 22;8:125. doi: 10.1186/1471-2164-8-125. BMC Genomics. 2007. PMID: 17519032 Free PMC article. - Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray.
Seki M, Narusaka M, Ishida J, Nanjo T, Fujita M, Oono Y, Kamiya A, Nakajima M, Enju A, Sakurai T, Satou M, Akiyama K, Taji T, Yamaguchi-Shinozaki K, Carninci P, Kawai J, Hayashizaki Y, Shinozaki K. Seki M, et al. Plant J. 2002 Aug;31(3):279-92. doi: 10.1046/j.1365-313x.2002.01359.x. Plant J. 2002. PMID: 12164808 - From laboratory to field. Using information from Arabidopsis to engineer salt, cold, and drought tolerance in crops.
Zhang JZ, Creelman RA, Zhu JK. Zhang JZ, et al. Plant Physiol. 2004 Jun;135(2):615-21. doi: 10.1104/pp.104.040295. Epub 2004 Jun 1. Plant Physiol. 2004. PMID: 15173567 Free PMC article. Review. No abstract available.
Cited by
- Genome wide transcriptional profile analysis of Vitis amurensis and Vitis vinifera in response to cold stress.
Xin H, Zhu W, Wang L, Xiang Y, Fang L, Li J, Sun X, Wang N, Londo JP, Li S. Xin H, et al. PLoS One. 2013;8(3):e58740. doi: 10.1371/journal.pone.0058740. Epub 2013 Mar 13. PLoS One. 2013. PMID: 23516547 Free PMC article. - Connecting the dots: from nanodomains to physiological functions of REMORINs.
Gouguet P, Gronnier J, Legrand A, Perraki A, Jolivet MD, Deroubaix AF, German-Retana S, Boudsocq M, Habenstein B, Mongrand S, Germain V. Gouguet P, et al. Plant Physiol. 2021 Apr 2;185(3):632-649. doi: 10.1093/plphys/kiaa063. Plant Physiol. 2021. PMID: 33793872 Free PMC article. Review. - Rates of evolution in stress-related genes are associated with habitat preference in two Cardamine lineages.
Ometto L, Li M, Bresadola L, Varotto C. Ometto L, et al. BMC Evol Biol. 2012 Jan 18;12:7. doi: 10.1186/1471-2148-12-7. BMC Evol Biol. 2012. PMID: 22257588 Free PMC article. - Obligate Biotroph Pathogens of the Genus Albugo Are Better Adapted to Active Host Defense Compared to Niche Competitors.
Ruhe J, Agler MT, Placzek A, Kramer K, Finkemeier I, Kemen EM. Ruhe J, et al. Front Plant Sci. 2016 Jun 20;7:820. doi: 10.3389/fpls.2016.00820. eCollection 2016. Front Plant Sci. 2016. PMID: 27379119 Free PMC article. - Expression patterns of genes involved in the defense and stress response of Spiroplasma citri infected Madagascar Periwinkle Catharanthus roseus.
Nejat N, Vadamalai G, Dickinson M. Nejat N, et al. Int J Mol Sci. 2012;13(2):2301-2313. doi: 10.3390/ijms13022301. Epub 2012 Feb 21. Int J Mol Sci. 2012. PMID: 22408455 Free PMC article.
References
- Adamska I, Kloppstech K. Low temperature increases the abundance of early light-inducible transcript under light stress conditions. J Biol Chem. 1994;269:30221–30226. - PubMed
- Adamska I, Roobol-Boza M, Lindahl M, Andersson B. Isolation of pigment-binding early light-inducible proteins from pea. Eur J Biochem. 1999;260:453–460. - PubMed
- Allen GJ, Chu SP, Schumacher K, Shimazaki CT, Vafeados D, Kemper A, Hawke SD, Tallman G, Tsien RY, Harper JF et al. Alteration of stimulus-specific guard cell calcium oscillations and stomatal closing in Arabidopsis det3 mutant. Science. 2000;289:2338–2342. - PubMed
- Apse MP, Aharon GS, Snedden WA, Blumwald E. Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science. 1999;285:1256–1258. - PubMed
- Blumwald E. Sodium transport and salt tolerance in plants. Curr Opin Cell Biol. 2000;12:431–434. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases