Transcriptional modulation of bacterial gene expression by subinhibitory concentrations of antibiotics - PubMed (original) (raw)
Transcriptional modulation of bacterial gene expression by subinhibitory concentrations of antibiotics
Ee-Been Goh et al. Proc Natl Acad Sci U S A. 2002.
Abstract
Antibiotics such as erythromycin and rifampicin, at low concentrations, alter global bacterial transcription patterns as measured by the stimulation or inhibition of a variety of promoter-lux reporter constructs in a Salmonella typhimurium library. Analysis of a 6,500-clone library indicated that as many as 5% of the promoters may be affected, comprising genes for a variety of functions, as well as a significant fraction of genes with no known function. Studies of a selection of the reporter clones showed that stimulation varied depending on the nature of the antibiotic, the promoter, and what culture medium was used; the response differed on solid as compared with liquid media. Transcription was markedly reduced in antibiotic-resistant hosts, but the presence of mutations deficient in stress responses such as SOS or universal stress did not prevent antibiotic-induced modulation. The results show that small molecules may have contrasting effects on bacteria depending on their concentration: either the modulation of bacterial metabolism by altering transcription patterns or the inhibition of growth by the inhibition of specific target functions. Both activities could play important roles in the regulation of microbial communities. These studies indicate that the detection of pharmaceutically useful natural product inhibitors could be effectively achieved by measuring activation of transcription at low concentrations in high-throughput assays using appropriate bacterial promoter-reporter constructs.
Figures
Fig 1.
Comparison between growth inhibition and promoter activation (light production) by erythromycin (A and B) and rifampicin (C and D) over a range of different concentrations as measured with Etest strips placed on bacterial cell overlays. (B and D) Luminescence. The _lux_–reporter constructions used are listed in Table 1. The bottom of the inhibition zone (where it intersects with the Etest strip) gives a measure of the MICs, which are 32 μg/ml (erythromycin) and 12 μg/ml (rifampicin).
Fig 2.
Combined scatter plot of the actions of rifampicin at 1 μg/ml and erythromycin at 5 μg/ml determined by using a 6,500-clone S. typhimurium random promoter–lux library. RLU, relative light units. Incubation in microtiter plate liquid cultures was for 24 h at 37°C. Points above the diagonal indicate promoter-activated strains and points below the diagonal indicate clones in which promoter activity was repressed. (-, erythromycin; ♦, rifampicin.)
Fig 3.
(Left) Scatter plots of the reassay of selected clones activated by erythromycin at 1 μg/ml (-) and 30 μg/ml (•). (Right) Reassay of selected clones activated by rifampicin at 0.2 μg/ml (×) and 2.5 μg/ml (•). CPS, counts per second. OD is at 620 nm; 1.E+01 = 101, etc.
Fig 4.
The effect of mutations to resistance to erythromycin (rplV) and to rifampicin (rpoB) in S. typhimurium on antibiotic inhibition and promoter activation on solid media. (A) Rifampicin-sensitive. (B) Rifampicin-resistant. (C) Erythromycin-sensitive. (D) Erythromycin-resistant.
Fig 5.
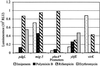
The antibiotic specificity of the transcription activation of different promoters. Overlays of S. typhimurium promoter–lux fusions on LB agar were exposed to discs containing antibiotics (10 μg of imipenem, 10 μg of polymixin B, 10 μg of rifampicin, or 15 μg of erythromycin). Light production was measured as described in Materials and Methods. RLU, relative light units. pheA* is a presumptive identification.
Fig 6.
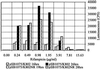
Concentration dependence of promoter activation (_plasI_′:luxCDABE) in a rifampicin-sensitive (K802; MIC 12 μg/ml) and a rifampicin-resistant (K802NR; MIC >256 μg/ml) E. coli host.
Similar articles
- Dual effects of MLS antibiotics: transcriptional modulation and interactions on the ribosome.
Tsui WH, Yim G, Wang HH, McClure JE, Surette MG, Davies J. Tsui WH, et al. Chem Biol. 2004 Sep;11(9):1307-16. doi: 10.1016/j.chembiol.2004.07.010. Chem Biol. 2004. PMID: 15380191 - Transcription modulation of Salmonella enterica serovar Typhimurium promoters by sub-MIC levels of rifampin.
Yim G, de la Cruz F, Spiegelman GB, Davies J. Yim G, et al. J Bacteriol. 2006 Nov;188(22):7988-91. doi: 10.1128/JB.00791-06. Epub 2006 Sep 15. J Bacteriol. 2006. PMID: 16980465 Free PMC article. - Modulation of Salmonella gene expression by subinhibitory concentrations of quinolones.
Yim G, McClure J, Surette MG, Davies JE. Yim G, et al. J Antibiot (Tokyo). 2011 Jan;64(1):73-8. doi: 10.1038/ja.2010.137. Epub 2010 Nov 24. J Antibiot (Tokyo). 2011. PMID: 21102598 - The world of subinhibitory antibiotic concentrations.
Davies J, Spiegelman GB, Yim G. Davies J, et al. Curr Opin Microbiol. 2006 Oct;9(5):445-53. doi: 10.1016/j.mib.2006.08.006. Epub 2006 Aug 30. Curr Opin Microbiol. 2006. PMID: 16942902 Review. - Antibiotic-supersusceptible mutants of Escherichia coli and Salmonella typhimurium.
Vaara M. Vaara M. Antimicrob Agents Chemother. 1993 Nov;37(11):2255-60. doi: 10.1128/AAC.37.11.2255. Antimicrob Agents Chemother. 1993. PMID: 8285603 Free PMC article. Review. No abstract available.
Cited by
- Subinhibitory antibiotic therapy alters recurrent urinary tract infection pathogenesis through modulation of bacterial virulence and host immunity.
Goneau LW, Hannan TJ, MacPhee RA, Schwartz DJ, Macklaim JM, Gloor GB, Razvi H, Reid G, Hultgren SJ, Burton JP. Goneau LW, et al. mBio. 2015 Mar 31;6(2):e00356-15. doi: 10.1128/mBio.00356-15. mBio. 2015. PMID: 25827417 Free PMC article. - Global transcriptional responses to the bacteriocin colicin M in Escherichia coli.
Kamenšek S, Žgur-Bertok D. Kamenšek S, et al. BMC Microbiol. 2013 Feb 19;13:42. doi: 10.1186/1471-2180-13-42. BMC Microbiol. 2013. PMID: 23421615 Free PMC article. - Sub-Optimal Treatment of Bacterial Biofilms.
Song T, Duperthuy M, Wai SN. Song T, et al. Antibiotics (Basel). 2016 Jun 22;5(2):23. doi: 10.3390/antibiotics5020023. Antibiotics (Basel). 2016. PMID: 27338489 Free PMC article. Review. - Antibiotic dialogues: induction of silent biosynthetic gene clusters by exogenous small molecules.
Okada BK, Seyedsayamdost MR. Okada BK, et al. FEMS Microbiol Rev. 2017 Jan;41(1):19-33. doi: 10.1093/femsre/fuw035. Epub 2016 Aug 29. FEMS Microbiol Rev. 2017. PMID: 27576366 Free PMC article. Review. - Chloramphenicol and tetracycline decrease motility and increase invasion and attachment gene expression in specific isolates of multidrug-resistant Salmonella enterica serovar Typhimurium.
Brunelle BW, Bearson BL, Bearson SM. Brunelle BW, et al. Front Microbiol. 2015 Jan 30;5:801. doi: 10.3389/fmicb.2014.00801. eCollection 2014. Front Microbiol. 2015. PMID: 25688233 Free PMC article.
References
- Waksman S. A. (1961) Perspect. Biol. Med. 4, 271-286.
- Kell D. B., Kaprelyants, A. S. & Grafen, A. (1995) Trends Ecol. Evol. 10, 126-129. - PubMed
- Demain A. L. & Fang, A. (2000) in History of Modern Biotechnology, ed. Fiechter, A. (Springer, Berlin), Vol. 1, pp. 2–39.
- Piepersberg W. (2001) in Molecular Medical Microbiology, ed. Sussman, M. (Academic, New York), Vol. 1, pp. 561–585.
- Dunny G. M. & Winans, S. C., (1999) Cell–Cell Signaling in Bacteria (Am. Soc. Microbiol., Washington, DC).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical