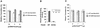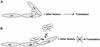The transformation suppressor Pdcd4 is a novel eukaryotic translation initiation factor 4A binding protein that inhibits translation - PubMed (original) (raw)
The transformation suppressor Pdcd4 is a novel eukaryotic translation initiation factor 4A binding protein that inhibits translation
Hsin-Sheng Yang et al. Mol Cell Biol. 2003 Jan.
Abstract
Pdcd4 is a novel transformation suppressor that inhibits tumor promoter-induced neoplastic transformation and the activation of AP-1-dependent transcription required for transformation. A yeast two-hybrid analysis revealed that Pdcd4 associates with the eukaryotic translation initiation factors eIF4AI and eIF4AII. Immunofluorescent confocal microscopy showed that Pdcd4 colocalizes with eIF4A in the cytoplasm. eIF4A is an ATP-dependent RNA helicase needed to unwind 5' mRNA secondary structure. Recombinant Pdcd4 specifically inhibited the helicase activity of eIF4A and eIF4F. In vivo translation assays showed that Pdcd4 inhibited cap-dependent but not internal ribosome entry site (IRES)-dependent translation. In contrast, Pdcd4(D418A), a mutant inactivated for binding to eIF4A, failed to inhibit cap-dependent or IRES-dependent translation or AP-1 transactivation. Recombinant Pdcd4 prevented eIF4A from binding to the C-terminal region of eIF4G (amino acids 1040 to 1560) but not to the middle region of eIF4G(amino acids 635 to 1039). In addition, both Pdcd4 and Pdcd4(D418A) bound to the middle region of eIF4G. The mechanism by which Pdcd4 inhibits translation thus appears to involve inhibition of eIF4A helicase, interference with eIF4A association-dissociation from eIF4G, and inhibition of eIF4A binding to the C-terminal domain of eIF4G. Pdcd4 binding to eIF4A is linked to its transformation-suppressing activity, as Pdcd4-eIF4A binding and consequent inhibition of translation are required for Pdcd4 transrepression of AP-1.
Figures
FIG. 1.
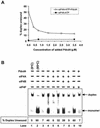
Identification of Pdcd4 binding to eIF4A. (A) Coimmunoprecipitation of Pdcd4 with eIF4A. JB6 P+ cell lysates isolated following transient transfection with Pdcd4 expression plasmid were immunoprecipitated (IP) with goat serum (lane 2) or eIF4A antibody (Ab) (lane 3). The immunoprecipitates were resolved by SDS-10% PAGE followed by immunoblotting with Pdcd4 antibody. Lane 1 shows one-tenth of the cell lysates. (B) Coomassie blue staining of GST-Pdcd4. One microgram of GST-Pdcd4 expressed in SF-9 cells and purified from a glutathione column as described in Materials and Methods was loaded onto SDS-PAGE (10%) and stained with SimpleBlue (Invitrogen). Lane M, protein molecular size markers. (C) GST pull-down of eIF4A with Pdcd4. JB6 P+ cell lysates isolated following transient transfection with Xpress-tagged eIF4A expression plasmid were pulled down with GST (lane 2) or GST-Pdcd4 (lane 3). The bound proteins were resolved by SDS-10% PAGE followed by immunoblotting with Xpress antibody. Lane 1 shows one-tenth of the cell lysates. (D) Mammalian two-hybrid assay of Pdcd4 binding to eIF4A. Various amounts (5, 10, and 50 ng) of plasmid pCMV-BD-Pdcd4 (or its empty vector, pCMV-DB) and pCMV-AD-eIF4A (or its empty vector, pCMV-AD) along with the Gal4-luciferase reporter gene were cotransfected into JB6 P+ cells. After 48 h, cells were lysed, and the luciferase activity was measured. The luciferase activity from the cells with 5 ng of pCMV-BD-Pdcd4 and 5 ng of pCMV-AD-eIF4A was designated as 1. These experiments were repeated three times, each with five independent transfections, and representative data are shown. Results are expressed as the mean ± standard deviation. RLU, relative luciferase units.
FIG. 2.
Immunofluorescent detection of colocalization of Pdcd4 and eIF4a in JB6 P− cells. P− cells were immunostained for Pdcd4 (A) and eIF4A (B) and viewed by confocal microscopy. (C) The merged images of panels A and B display a yellow color indicative of colocalization of the two proteins to the perinuclear region of the cytoplasm. The cell shown is a representative example of multiple P− cells. Of seven randomly selected cells, 63% ± 5% of total Pdcd4 was colocalized with the eIF4A, and 65% ± 6% of total eIF4A was colocalized with Pdcd4 in the cytoplasm. The nucleus, which did not show either Pdcd4 or eIF4A staining, was masked for the purpose of limiting the area and therefore the stringency by which the spatial statistical algorithm could test whether the colocalization was the result of random overlap. The estimated probability that the colocalization was due to random overlap was 0.1%. Bar, 15 μm.
FIG. 3.
Inhibition of eIF4A RNA helicase activity by Pdcd4. (A) Pdcd4 inhibits eIF4A helicase activity. Unwinding of a 2 nM RNA duplex (12 bp; Δ_G_ = −21.4 kcal/mol) by 1.5 μM eIF4A (open circles) was performed as described in Materials and Methods. As a control (solid squares), eIF4A was incubated with the duplex in the absence of ATP. Separate controls indicated that there was no unwinding by Pdcd4 in the presence of 1 mM ATP (not shown). After the 15-min incubation at 35°C, unwinding was quantitated by gel electrophoresis and subsequent analysis with an Ambis radioanalytic scanner. (B) Pdcd4 inhibits eIF4F helicase activity and eIF4B does not stimulate eIF4A helicase activity in the presence of Pdcd4. A 2 nM RNA duplex (13 bp; Δ_G_ = −23.1 kcal/mol) was incubated with the following proteins: 13.5 pmol of eIF4A (lane 4), 13.5 pmol of eIF4A plus 27 pmol of Pdcd4 (lane 5), 6.75 pmol of eIF4A (lane 6), 6.75 pmol of eIF4A plus 6.75 pmol of eIF4B (lane 7), 6.75 pmol of eIF4A plus 6.75 pmol of eIF4B plus 13.5 pmol of Pdcd4 (lane 8), 6.75 pmol of eIF4F (lane 9), or 6.75 pmol of eIF4F plus 13.5 pmol of Pdcd4 (lane 10) for 15 min at 35°C. Samples were separated and quantitated as described in Materials and Methods. Lane 1, duplex RNA incubated under unwinding conditions without protein for 15 min at 0°C; lane 2, duplex RNA incubated under the same conditions for 5 min at 95°C; lane 3, duplex RNA incubated under the same conditions without ATP and proteins for 15 min at 35°C. (C) Pdcd4 inhibits helicase activities of eIF4A plus eIF4B and eIF4F in a concentration-dependent manner. Unwinding of a 2 nM RNA duplex (13 bp; Δ_G_ = −23.1 kcal/mol) by eIF4A (6.75 pmol) plus eIF4B (6.75 pmol) without (lane 3) or with increasing concentrations of Pdcd4 (3.4 to 13.5 pmol, lanes 4 to 6) or by eIF4F (6.75 pmol) without (lane 7) or with increasing concentrations of Pdcd4 (3.4 pmol to 13.5 pmol, lanes 8 to 10) was performed as described in Materials and Methods. Lane 1, duplex RNA incubated under unwinding conditions without protein for 15 min at 0°C; lane 2, duplex RNA incubated under the same conditions for 5 min at 95°C. (D) Pdcd4 does not inhibit Ded1p helicase activity. Unwinding of a 2 nM RNA duplex (13 bp; Δ_G_ = −23.1 kcal/mol) by Ded1p helicase (0.5 pmol) in the absence (lane 4) or presence (lanes 5 and 6, 1 and 2 pmol, respectively) of Pdcd4 was performed as described in Materials and Methods. Lane 1, duplex RNA incubated under unwinding conditions without protein for 15 min at 0°C; lane 2, duplex RNA incubated under the same conditions for 5 min at 95°C.
FIG. 3.
Inhibition of eIF4A RNA helicase activity by Pdcd4. (A) Pdcd4 inhibits eIF4A helicase activity. Unwinding of a 2 nM RNA duplex (12 bp; Δ_G_ = −21.4 kcal/mol) by 1.5 μM eIF4A (open circles) was performed as described in Materials and Methods. As a control (solid squares), eIF4A was incubated with the duplex in the absence of ATP. Separate controls indicated that there was no unwinding by Pdcd4 in the presence of 1 mM ATP (not shown). After the 15-min incubation at 35°C, unwinding was quantitated by gel electrophoresis and subsequent analysis with an Ambis radioanalytic scanner. (B) Pdcd4 inhibits eIF4F helicase activity and eIF4B does not stimulate eIF4A helicase activity in the presence of Pdcd4. A 2 nM RNA duplex (13 bp; Δ_G_ = −23.1 kcal/mol) was incubated with the following proteins: 13.5 pmol of eIF4A (lane 4), 13.5 pmol of eIF4A plus 27 pmol of Pdcd4 (lane 5), 6.75 pmol of eIF4A (lane 6), 6.75 pmol of eIF4A plus 6.75 pmol of eIF4B (lane 7), 6.75 pmol of eIF4A plus 6.75 pmol of eIF4B plus 13.5 pmol of Pdcd4 (lane 8), 6.75 pmol of eIF4F (lane 9), or 6.75 pmol of eIF4F plus 13.5 pmol of Pdcd4 (lane 10) for 15 min at 35°C. Samples were separated and quantitated as described in Materials and Methods. Lane 1, duplex RNA incubated under unwinding conditions without protein for 15 min at 0°C; lane 2, duplex RNA incubated under the same conditions for 5 min at 95°C; lane 3, duplex RNA incubated under the same conditions without ATP and proteins for 15 min at 35°C. (C) Pdcd4 inhibits helicase activities of eIF4A plus eIF4B and eIF4F in a concentration-dependent manner. Unwinding of a 2 nM RNA duplex (13 bp; Δ_G_ = −23.1 kcal/mol) by eIF4A (6.75 pmol) plus eIF4B (6.75 pmol) without (lane 3) or with increasing concentrations of Pdcd4 (3.4 to 13.5 pmol, lanes 4 to 6) or by eIF4F (6.75 pmol) without (lane 7) or with increasing concentrations of Pdcd4 (3.4 pmol to 13.5 pmol, lanes 8 to 10) was performed as described in Materials and Methods. Lane 1, duplex RNA incubated under unwinding conditions without protein for 15 min at 0°C; lane 2, duplex RNA incubated under the same conditions for 5 min at 95°C. (D) Pdcd4 does not inhibit Ded1p helicase activity. Unwinding of a 2 nM RNA duplex (13 bp; Δ_G_ = −23.1 kcal/mol) by Ded1p helicase (0.5 pmol) in the absence (lane 4) or presence (lanes 5 and 6, 1 and 2 pmol, respectively) of Pdcd4 was performed as described in Materials and Methods. Lane 1, duplex RNA incubated under unwinding conditions without protein for 15 min at 0°C; lane 2, duplex RNA incubated under the same conditions for 5 min at 95°C.
FIG. 4.
Inhibition of in vitro translation by Pdcd4. (A) Rabbit reticulocyte lysate was preincubated with eIF4A alone (lane 1), with increasing amounts of Pdcd4 (0.15 to 4.8 μg, lanes 2 to 7), or with Pdcd4 and increasing amounts of eIF4A (lanes 8 to 10) for 5 min at 30°C prior to the addition of the bicistronic CAT/EMCV/LUC mRNA (0.2 μg). Translation was performed in a total volume of 20 μl as described in Materials and Methods. The band intensity was determined by Phosphorimager. The value obtained for both cap- and IRES-dependent translation in the absence of added Pdcd4 and eIF4A was designated as 100%. (B) Rabbit reticulocyte lysate was incubated with eIF4F (lanes 1 to 3) or eIF4F and Pdcd4 (lanes 4 to 6) for 5 min at 30°C prior to the addition of the bicistronic CAT/EMCV/LUC mRNA (0.2 μg). Translation was performed in a total volume of 20 μl as described in Materials and Methods. The band intensity was determined by Phosphorimager. The value obtained for both cap- and IRES-dependent translation in the absence of added Pdcd4 and eIF4F was designated as 100%.
FIG. 5.
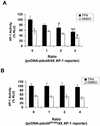
Pdcd4 inhibits translation in vivo. (A and C) The plasmid pcDNA-CAT/EMCV/LUC reporter system (0.2 μg) was transiently transfected with (A) pcDNA-Pdcd4 (0 to 2 μg) or (C) pcDNA-Pdcd4D418A (0 to 2 μg) into JB6 RT101 cells. Total DNA was maintained at 2.2 μg by adding pcDNA3.1+ vector DNA. After transfection, cells were serum starved (0.2% fetal bovine serum) for 24 h and then incubated with normal medium (4% fetal bovine serum) for an additional 24 h. The CAT and luciferase activities from the cells with 0 μg of pcDNA-Pdcd4 (A) or pcDNA-Pdcd4D418A (C) transfection was designated as 100%. These experiments were repeated three times in triplicate, and representative data are shown. Results are expressed as mean ± standard deviation. * and ** indicate significant differences compared with the control as determined by Student's t test (*, <0.005; **, <0.0001). (B) Pdcd4D418A mutant does not bind to eIF4A. Plasmid pCMV-BD-Pdcd4 (50 ng) (wild type [WT]) or pCMV-BD-Pdcd4D418A (50 ng) (D418A) was transiently transfected with pCMV-AD-eIF4A (50 ng) and Gal4-luciferase reporter DNA (25 ng) into JB6 RT101 cells. After 48 h, cells were lysed and luciferase activity was measured. The luciferase activity of wild-type Pdcd4 was designated as 100%. These experiments were repeated three times with five independent transfections each, and representative data are shown. Results are expressed as mean ± standard deviation. The inset shows an immunoblot of RT101 cells transiently transfected with pcDNA3.1+ (lane 1), pcDNA-Pdcd4 (lane 2), or pcDNA-Pdcd4D418A (lane 3) and detected with Pdcd4 antibody.
FIG. 6.
Pdcd4D418A does not inhibit AP-1-dependent transcription. JB6 P+ cells were transfected with 0.2 μg of the 4× AP-1 luciferase reporter gene and increasing amounts (0 to 0.8 μg) of pcDNA-Pdcd4 (A) or pcDNA-Pdcd4D418A (B). Total DNA was maintained at 1.0 μg by adding pcDNA3.1+ vector DNA. The luciferase activity of cells treated with tetradecanoyl phorbol acetate (TPA) and without pcDNA-Pdcd4 or pcDNA-Pdcd4D418A was designated as 100%. These experiments were repeated three times in triplicate, and representative data are shown. Results are expressed as mean ± standard deviation. * and ** indicate significant differences compared with the control (tetradecanoyl phorbol acetate or dimethyl sulfoxide [DMSO] treatment following transfection with 0 μg of pcDNA-Pdcd4) as determined by Student's t test (*, <0.005; **, <0.0001).
FIG. 7.
Prevention of eIF4A binding to the C-terminal but not to the middle one-third of eIF4G by Pdcd4: Pdcd4 binds to the middle one-third of eIF4G. (A) Structures of eIF4G1 and Pdcd4. The numbers refer to the size (in amino acids) of eIF4G1 and Pdcd4 and to the locations of the eIF4A binding domain and MA-3 domain (2, 32, 40). eIF4A binding domains (open box and arrows) in eIF4G1 are indicated schematically. The MA-3 domains (grey box) in eIF4G1 and Pdcd4 are indicated schematically. (B) Coomassie blue staining of recombinant GST-eIF4G(672-1065), GST-eIF4G(1201-1445), His-eIF4A, and His-Pdcd4. Three micrograms of each recombinant GST-eIF4G(672-1065) (lane 2), GST-eIF4G(1201-1445) (lane 3), His-Pdcd4 (lane 4), and His-eIF4A (lane 5) was resolved by SDS-PAGE and stained with SimpleBlue (Invitrogen). Lane 1, proteinmolecular size markers. (C) In vitro binding assay. Bovine liver GST (lanes 1 and 5), recombinant GST-eIF4G(1201-1445) (lanes 2 to 4), or GST-eIF4G(672-1065) (lanes 6 to 8) was immobilized on glutathione-Sepharose beads and incubated with 5 μg of His-Pdcd4 only (lanes 2 and 6), 5 μg of His-eIF4A only (lanes 3 and 7), or 5 μg of both His-Pdcd4 and His-eIF4A (lanes 1, 4, 5, and 8) on ice for 10 min. After being washed with binding buffer, the bound proteins were resolved by SDS-PAGE and analyzed by immunoblotting with GST antibody (first panel), penta-His antibody (second panel), or Pdcd4 antibody (third panel). Ten percent of input His-Pdcd4 and His-eIF4A proteins were subjected to SDS-PAGE followed by immunoblotting with penta-His antibody (fourth panel). GST-eIF4G(672-1065) and GST-eIF4G(1201-1445) immobilized on glutathione-Sepharose beads were shown as similar amounts. (D) Pulldown of Pdcd4 and Pdcd4D418A with GST-eIF4G(627-1065). JB6 P+ cell lysates isolated following transient transfection with pcDNA3.1+ (lanes 1 to 3), pcDNA-Pdcd4 (lanes 4 to 6), or pcDNA-Pdcd4D418A (lanes 7 to 9) were pulled down with GST (lanes 2, 5, and 8) or GST-eIF4G(627-1065) (lanes 3, 6, and 9). The bound proteins were resolved by SDS-10% PAGE followed by immunoblotting with Pdcd4 or eIF4A antibodies. (E) GST pulldown of endogenous eIF4G with Pdcd4. JB6 P+ cell lysate was pulled down with GST (lane 2) or GST-Pdcd4 (lane 3). The bound proteins were resolved by SDS-10% PAGE followed by immunoblotting with eIF4G antibody. Lane 1 shows one-tenth of the cell lysate.
FIG. 8.
Model of how Pdcd4 inhibits translation. (A) A sandwich model of eIF4A binding to eIF4G is shown (32). The eIF4A molecule binds to the middle and C-terminal one-third of eIF4G. (B) Model for Pdcd4 inhibition of translation. Pdcd4 inhibits the helicase activity of eIF4A. This inactivated eIF4A molecule is further trapped by Pdcd4 on the middle one-third of eIF4G. This process will block eIF4A association-dissociation cycling and keep the eIF4F complex inactivated. In addition, Pdcd4 also prevents from eIF4A binding to the C-terminal one-third of eIF4G. This will prevent eIF4A from stimulating the activity of eIF4F.
Similar articles
- A novel function of the MA-3 domains in transformation and translation suppressor Pdcd4 is essential for its binding to eukaryotic translation initiation factor 4A.
Yang HS, Cho MH, Zakowicz H, Hegamyer G, Sonenberg N, Colburn NH. Yang HS, et al. Mol Cell Biol. 2004 May;24(9):3894-906. doi: 10.1128/MCB.24.9.3894-3906.2004. Mol Cell Biol. 2004. PMID: 15082783 Free PMC article. - Mutational analysis of the DEAD-box RNA helicase eIF4AII characterizes its interaction with transformation suppressor Pdcd4 and eIF4GI.
Zakowicz H, Yang HS, Stark C, Wlodawer A, Laronde-Leblanc N, Colburn NH. Zakowicz H, et al. RNA. 2005 Mar;11(3):261-74. doi: 10.1261/rna.7191905. Epub 2005 Jan 20. RNA. 2005. PMID: 15661843 Free PMC article. - Structural basis for translational inhibition by the tumour suppressor Pdcd4.
Loh PG, Yang HS, Walsh MA, Wang Q, Wang X, Cheng Z, Liu D, Song H. Loh PG, et al. EMBO J. 2009 Feb 4;28(3):274-85. doi: 10.1038/emboj.2008.278. Epub 2009 Jan 15. EMBO J. 2009. PMID: 19153607 Free PMC article. - Manipulation of the host translation initiation complex eIF4F by DNA viruses.
Walsh D. Walsh D. Biochem Soc Trans. 2010 Dec;38(6):1511-6. doi: 10.1042/BST0381511. Biochem Soc Trans. 2010. PMID: 21118117 Review. - The Impact of Pdcd4, a Translation Inhibitor, on Drug Resistance.
Wang Q, Yang HS. Wang Q, et al. Pharmaceuticals (Basel). 2024 Oct 19;17(10):1396. doi: 10.3390/ph17101396. Pharmaceuticals (Basel). 2024. PMID: 39459035 Free PMC article. Review.
Cited by
- Exosomal microRNAs: Pleiotropic Impacts on Breast Cancer Metastasis and Their Clinical Perspectives.
Tang LB, Ma SX, Chen ZH, Huang QY, Wu LY, Wang Y, Zhao RC, Xiong LX. Tang LB, et al. Biology (Basel). 2021 Apr 7;10(4):307. doi: 10.3390/biology10040307. Biology (Basel). 2021. PMID: 33917233 Free PMC article. Review. - Ubiquitin ligases in cancer: Functions and clinical potentials.
Duan S, Pagano M. Duan S, et al. Cell Chem Biol. 2021 Jul 15;28(7):918-933. doi: 10.1016/j.chembiol.2021.04.008. Epub 2021 May 10. Cell Chem Biol. 2021. PMID: 33974914 Free PMC article. Review. - Tumour suppressive function and modulation of programmed cell death 4 (PDCD4) in ovarian cancer.
Wei N, Liu SS, Chan KK, Ngan HY. Wei N, et al. PLoS One. 2012;7(1):e30311. doi: 10.1371/journal.pone.0030311. Epub 2012 Jan 17. PLoS One. 2012. PMID: 22272332 Free PMC article. - The mechanistic insight of a specific interaction between 15d-Prostaglandin-J2 and eIF4A suggests an evolutionary conserved role across species.
Yun SJ, Kim H, Jung SH, Kim JH, Ryu JE, Singh NJ, Jeon J, Han JK, Kim CH, Kim S, Jang SK, Kim WJ. Yun SJ, et al. Biol Open. 2018 Nov 13;7(11):bio035402. doi: 10.1242/bio.035402. Biol Open. 2018. PMID: 30257829 Free PMC article. - mRNA Translation Is Dynamically Regulated to Instruct Stem Cell Fate.
Wang R, Amoyel M. Wang R, et al. Front Mol Biosci. 2022 Mar 31;9:863885. doi: 10.3389/fmolb.2022.863885. eCollection 2022. Front Mol Biosci. 2022. PMID: 35433828 Free PMC article. Review.
References
- Abramson, R. D., T. E. Dever, T. G. Lawson, B. K. Ray, R. E. Thach, and W. C. Merrick. 1987. The ATP−dependent interaction of eukaryotic initiation factors with mRNA. J. Biol. Chem. 262:3826-3832. - PubMed
- Bauer, C., I. Diesinger, N. Brass, H. Steinhart, H. Iro, and E. U. Meese. 2001. Translation initiation factor eIF-4G is immunogenic, overexpressed, and amplified in patients with squamous cell lung carcinoma. Cancer 92:822-829. - PubMed
- Brown, E. J., and S. L. Schreiber. 1996. A signaling pathway to translational control. Cell 86:517-520. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous