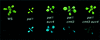An Arabidopsis SET domain protein required for maintenance but not establishment of DNA methylation - PubMed (original) (raw)
An Arabidopsis SET domain protein required for maintenance but not establishment of DNA methylation
Fabienne Malagnac et al. EMBO J. 2002.
Abstract
Cytosine methylation is critical for correct development and genome stability in mammals and plants. In order to elucidate the factors that control genomic DNA methylation patterning, a genetic screen for mutations that disrupt methylation-correlated silencing of the endogenous gene PAI2 was conducted in Arabidopsis: This screen yielded seven loss-of-function alleles in a SET domain protein with histone H3 Lys9 methyltransferase activity, SUVH4. The mutations conferred reduced cytosine methylation on PAI2, especially in non-CG sequence contexts, but did not affect methylation on another PAI locus carrying two genes arranged as an inverted repeat. Moreover, an unmethylated PAI2 gene could be methylated de novo in the suvh4 mutant background. These results suggest that SUVH4 is involved in maintenance but not establishment of methylation at particular genomic regions. In contrast, a heterochromatin protein 1 homolog, LHP1, had no effect on PAI methylation.
Figures
Fig. 1. PAI2 silencing is suppressed by suvh4 mutations. Representative 2.5-week-old plants of the indicated strains are shown under visible (upper panel) and UV (lower panel) light. The suvh4302* and cmt3i11a alleles were used. The suppressed phenotype of suvh4R302* is similar to those of the other six suvh4 alleles.
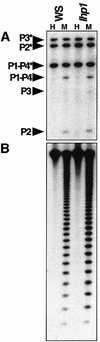
Fig. 2. suvh4 mutations confer reduced PAI2 methylation. (A) The _Hpa_II (H) and _Msp_I (M) restriction map of each WS PAI locus is shown, with the probed regions indicated by gray bars and the bisulfite sequenced regions indicated by hatched bars. (B) Genomic DNAs prepared from 4-week-old plants of the indicated genotypes were cleaved with either _Hpa_II or _Msp_I and used for Southern blot analysis with a PAI probe. P1-P4 is PAI1–PAI4, P2 is PAI2, and P3 is PAI3, with asterisks indicating the positions of species methylated at internal _Hpa_II/_Msp_I sites. Note that PAI3 is divergent from the probe sequence and thus gives a weaker signal than other PAI genes. (C) The blot shown in (B) was reprobed with a 180 bp CEN repeat probe. The phenotypes of the WS suvh4R302* allele shown are representative of the phenotypes observed with six other suhv4 alleles. The phenotypes of the WS pai1 suvh4R302* cmt3i11a double mutant line shown are representative of the phenotypes observed with three other independent double mutant lines.
Fig. 3. Bisulfite sequencing analysis of the PAI genes in a suvh4 mutant background. (A) Bisulfite genomic methylation sequencing was performed for the top strands of the PAI1 and PAI2 upstream regions in WS pai1 suvh4R302* or WS pai1 suvh4R302* cmt3i11a DNA. For each region, eight independent molecules were sequenced. Vertical lines indicate positions of cytosines, with the height of each line representing how many sequenced molecules had 5-methylcytosine (5-Me-C). Black indicates CG cytosines, blue indicates CNG cytosines, and red indicates other cytosines. Asterisks indicate sites with no methylation. The black horizontal line indicates the region of PAI identity, and the gray horizontal line indicates flanking upstream heterologous sequence unique to each gene. The exon and intron structures of PAI1 and PAI2 are shown as open boxes and dashed lines, respectively, under each sequence. These structures are based on full-length cDNA sequences for each gene (Melquist et al., 1999). (B) Within the region of PAI identity (black horizontal line in A), the percentages of available cytosines that are methylated in CG (black bars), CNG (white bars) or other (gray bars) sequence contexts are shown for either PAI1 (P1) or PAI2 (P2) in the indicated strains. Data for wild-type WS and WS pai1 cmt3 are from previous publications (Luff et al., 1999; Bartee et al., 2001). See also Supplementary figure 2.
Fig. 4. Positions of seven suvh4 alleles recovered as PAI2 silencing suppressors. The predicted amino acid sequence of WS SUVH4 is shown, with the positions of introns marked with black arrowheads. Missense mutations are indicated by the new amino acid above the affected codon. Nonsense mutations are indicated by an asterisk above the affected codon. Splice mutations are indicated by an x at the appropriate junction of the affected intron. The YDG, pre-SET, SET and post-SET domains as previously defined for SUVH4 relative to related proteins (Baumbusch et al., 2001; Jackson et al., 2002) are underlined. The WS genomic sequence of SUVH4 is available as DDBJ/EMBL/GenBank accession No. AF538715.
Fig. 5. A SUVH4 transgene can complement the suvh4 mutant defect. (A) Representative T2 2-week-old seedlings of the suvh4R302* mutant transformed with either a SUVH4 genomic clone or a CMT3 genomic clone (two independent lines each) are shown under visible (upper panel) or UV (lower panel) light. (B) Genomic DNAs prepared from single leaves of 4-week-old T2 plants of the same lines shown in (A) were cleaved with either _Hpa_II (H) or _Msp_I (M) and used for Southern blot analysis with a PAI probe. P1-P4 is PAI1–PAI4, P2 is PAI2, and P3 is PAI3, with asterisks indicating the positions of species methylated at internal _Hpa_II/_Msp_I sites.
Fig. 6. The lhp1 mutation does not affect PAI or CEN methylation in WS. (A) Genomic DNAs prepared from 4-week-old plants of the indicated genotypes were cleaved with either _Hpa_II (H) or _Msp_I (M) and used for Southern blot analysis with a PAI probe. P1-P4 is PAI1–PAI4, P2 is PAI2, and P3 is PAI3, with asterisks indicating the positions of species methylated at internal _Hpa_II/_Msp_I sites. (B) The blot shown in (A) was reprobed with a 180 bp CEN repeat probe. The lhp1-1 allele, which displays characteristic developmental defects, was used for this analysis (Gaudin et al., 2001).
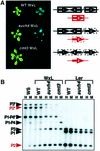
Fig. 7. The WS pai1–PAI4 inverted repeat locus triggers de novo methylation and silencing of an unmethylated Ler PAI2 gene in a SUVH4-deficient but not a CMT3-deficient background. (A) To the left, representative WxL wild-type (F4), WxL suvh4 (F5) or WxL cmt3 (F5) 2.5-week-old seedlings are shown under visible (left panel) or UV (right panel) light. To the right, diagrams show the PAI genotypes and methylation phenotypes in these lines. PAI genes inherited from WS are indicated as black arrows. Note that none of the three WS PAI genes encodes functional PAI enzyme. The WS PAI1–PAI4 and PAI3 loci both lie on the upper arm of chromosome 1, indicated by a slash separating the two loci. The Ler PAI2 gene is indicated by a red arrow. Methylation is indicated by boxes around the affected genes, with a solid line representing dense methylation and a dashed line representing partial methylation. (B) Genomic DNAs prepared from 4-week-old plants of the indicated strains were cleaved with either _Hpa_II (H) or _Msp_I (M) and used for Southern blot analysis with a PAI probe. P1-P4 is WS PAI1–PAI4, P1 is Ler PAI1, P2 is PAI2, and P3 is PAI3, with asterisks indicating the positions of species methylated at internal _Hpa_II/_Msp_I sites. WxL wild-type, WxL suvh4 and WxL cmt3 DNAs were prepared from F4, F5 and F5 generation plants, respectively.
Similar articles
- H3 lysine 9 methylation is maintained on a transcribed inverted repeat by combined action of SUVH6 and SUVH4 methyltransferases.
Ebbs ML, Bartee L, Bender J. Ebbs ML, et al. Mol Cell Biol. 2005 Dec;25(23):10507-15. doi: 10.1128/MCB.25.23.10507-10515.2005. Mol Cell Biol. 2005. PMID: 16287862 Free PMC article. - The SRA methyl-cytosine-binding domain links DNA and histone methylation.
Johnson LM, Bostick M, Zhang X, Kraft E, Henderson I, Callis J, Jacobsen SE. Johnson LM, et al. Curr Biol. 2007 Feb 20;17(4):379-84. doi: 10.1016/j.cub.2007.01.009. Epub 2007 Jan 18. Curr Biol. 2007. PMID: 17239600 Free PMC article. - Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase.
Jackson JP, Lindroth AM, Cao X, Jacobsen SE. Jackson JP, et al. Nature. 2002 Apr 4;416(6880):556-60. doi: 10.1038/nature731. Epub 2002 Mar 17. Nature. 2002. PMID: 11898023 - Heterochromatin proteins and the control of heterochromatic gene silencing in Arabidopsis.
Fischer A, Hofmann I, Naumann K, Reuter G. Fischer A, et al. J Plant Physiol. 2006 Feb;163(3):358-68. doi: 10.1016/j.jplph.2005.10.015. Epub 2005 Dec 27. J Plant Physiol. 2006. PMID: 16384625 Review. - The many faces of histone lysine methylation.
Lachner M, Jenuwein T. Lachner M, et al. Curr Opin Cell Biol. 2002 Jun;14(3):286-98. doi: 10.1016/s0955-0674(02)00335-6. Curr Opin Cell Biol. 2002. PMID: 12067650 Review.
Cited by
- The SET-domain protein SUVR5 mediates H3K9me2 deposition and silencing at stimulus response genes in a DNA methylation-independent manner.
Caro E, Stroud H, Greenberg MV, Bernatavichute YV, Feng S, Groth M, Vashisht AA, Wohlschlegel J, Jacobsen SE. Caro E, et al. PLoS Genet. 2012;8(10):e1002995. doi: 10.1371/journal.pgen.1002995. Epub 2012 Oct 11. PLoS Genet. 2012. PMID: 23071452 Free PMC article. - Epimutations are associated with CHROMOMETHYLASE 3-induced de novo DNA methylation.
Wendte JM, Zhang Y, Ji L, Shi X, Hazarika RR, Shahryary Y, Johannes F, Schmitz RJ. Wendte JM, et al. Elife. 2019 Jul 29;8:e47891. doi: 10.7554/eLife.47891. Elife. 2019. PMID: 31356150 Free PMC article. - Mass spectrometry analysis of Arabidopsis histone H3 reveals distinct combinations of post-translational modifications.
Johnson L, Mollah S, Garcia BA, Muratore TL, Shabanowitz J, Hunt DF, Jacobsen SE. Johnson L, et al. Nucleic Acids Res. 2004 Dec 14;32(22):6511-8. doi: 10.1093/nar/gkh992. Print 2004. Nucleic Acids Res. 2004. PMID: 15598823 Free PMC article. - Major transcriptomic differences are induced by warmer temperature conditions experienced during asexual and sexual reproduction in Fragaria vesca ecotypes.
Zhang Y, Viejo M, Yakovlev I, Tengs T, Krokene P, Hytönen T, Grini PE, Fossdal CG. Zhang Y, et al. Front Plant Sci. 2023 Jul 14;14:1213311. doi: 10.3389/fpls.2023.1213311. eCollection 2023. Front Plant Sci. 2023. PMID: 37521931 Free PMC article. - Histone modifications and chromatin dynamics: a focus on filamentous fungi.
Brosch G, Loidl P, Graessle S. Brosch G, et al. FEMS Microbiol Rev. 2008 May;32(3):409-39. doi: 10.1111/j.1574-6976.2007.00100.x. Epub 2008 Jan 23. FEMS Microbiol Rev. 2008. PMID: 18221488 Free PMC article. Review.
References
- Baumbusch L.O., Thorstensen,T., Krauss,V., Fischer,A., Naumann,K., Assalkhou,R., Schulz,I., Reuter,G. and Aalen,R.B. (2001) The Arabidopsis thaliana genome contains at least 29 active genes encoding SET domain proteins that can be assigned to four evolutionarily conserved classes. Nucleic Acids Res., 29, 4319–4333. - PMC - PubMed
- Bell C.J. and Ecker,J.R. (1994) Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics, 19, 137–144. - PubMed
- Bender J. (2001) A vicious cycle: RNA silencing and DNA methylation in plants. Cell, 106, 129–132. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials