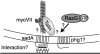SadA, a novel adhesion receptor in Dictyostelium - PubMed (original) (raw)
SadA, a novel adhesion receptor in Dictyostelium
Petra Fey et al. J Cell Biol. 2002.
Abstract
Little is known about cell-substrate adhesion and how motile and adhesive forces work together in moving cells. The ability to rapidly screen a large number of insertional mutants prompted us to perform a genetic screen in Dictyostelium to isolate adhesion-deficient mutants. The resulting substrate adhesion-deficient (sad) mutants grew in plastic dishes without attaching to the substrate. The cells were often larger than their wild-type parents and displayed a rough surface with many apparent blebs. One of these mutants, sadA-, completely lacked substrate adhesion in growth medium. The sadA- mutant also showed slightly impaired cytokinesis, an aberrant F-actin organization, and a phagocytosis defect. Deletion of the sadA gene by homologous recombination recreated the original mutant phenotype. Expression of sadA-GFP in sadA-null cells restored the wild-type phenotype. In sadA-GFP-rescued mutant cells, sadA-GFP localized to the cell surface, appropriate for an adhesion molecule. SadA contains nine putative transmembrane domains and three conserved EGF-like repeats in a predicted extracellular domain. The EGF repeats are similar to corresponding regions in proteins known to be involved in adhesion, such as tenascins and integrins. Our data combined suggest that sadA is the first substrate adhesion receptor to be identified in Dictyostelium.
Figures
Figure 1.
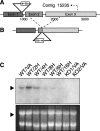
SadA locus and transcript. (A) SadA gene structure and plasmid insertion site. In REMI mutant 3IIG11, the transforming plasmid pUCΔBamBsr inserted at the 3′ end of the first exon, disrupting transcription of the remainder of the gene. Indicated at position 2790 is where the sequence of contig 15235 ends. (B) Structure of the knock-out construct. The plasmid pUCΔBamBsr containing genomic flanking sequences was inserted by homologous recombination so that the vector inserted in opposite direction as compared with the initial REMI mutant and resulted in the deletion of a 1-kb genomic sequence. (C) Northern blot analysis. In vegetative wild-type cells (WT/VA), the sadA transcript is ∼3.5 kb long. The presence of the transcript in developing wild type was tested after 2, 4, 6, 8, and 16 h starvation. Note that the gene is significantly down-regulated after 4 h starvation. The sadA transcript is absent in vegetative amoebae of two independent knock-out strains (KO1/VA and KO2/VA). As a loading control, the ethidium bromide–stained gel is shown.
Figure 2.
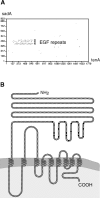
Similarity of sadA to tenascin A (chicken) and predicted protein organization. (A) The dot matrix alignment illustrates the high similarity of sadA's three and tenascin A's 13.5 EGF-like repeats. There is virtually no similarity outside this region. The dot matrix plot was created by the Vector NTI Suite 6 (Informax Inc.), using a window size of 30 amino acids and a stringency of 30%. (B) According to the prediction of the TMHMM V2.0 computer program, nine transmembrane regions are depicted. The 25 amino acids predicted to be a signal peptide (SignalP V1.1) were omitted in this model. Each EGF-like repeat comprises six conserved cysteines (black dot) and one conserved glycine residue (dark gray dot).
Figure 3.
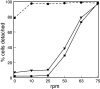
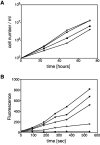
SadA-null cells cannot initiate attachment. Cells were plated at 105 per ml, grown overnight, and subjected to rotation on an orbital shaker. Subsequently, cells floating in the medium were counted. In the 0-rpm data, the number of detached cells was determined without prior agitation. Here, because most sadA-null cells (closed circles) sank to the surface by gravity overnight, recovery was only ∼80%, not close to 100%, although the cells were generally not attached. In all other samples, the detached cells were counted after they were subjected to shear stress from 10 to 75 rpm for 1 h. After agitation at any speed, nearly all sadA-null cells were in the supernatant. In comparison, even at 50 rpm, 71% of wild-type cells (closed squares), and 61% of sadA–GFP-rescued cells (closed inverted triangles) remained attached. Only vigorous shaking at 75 rpm detached all wild-type and rescued cells.
Figure 4.
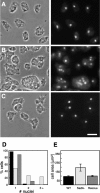
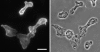
SadA-null cells are generally larger and multinucleate. (A) Wild-type AX3, (B) sadA-null, and (C) sadA–GFP-rescued cells. Cells were flattened under an agarose sheet before fixation. Note the correlation of cell size and nuclei number. Bar, 10 μm. (D) A graph illustrating the nuclei number of wild type (black bar), sadA null (light gray), and sadA–GFP rescue (dark gray). For each cell type, the nuclei of 200 cells were counted. (E) A graph illustrating the cell surface area. 12 fixed and flattened cells were measured for each cell type.
Figure 5.
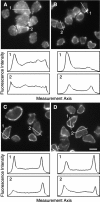
F-actin is mislocalized in vegetative and indistuinguishable from wild type in starved cells. Actin was stained with Alexa®568, and actin intensity profiles were created from cross sections of two cells each. (A) Vegetative sadA-null cells. (B) Vegetative wild-type cells. (C) 4-h developed sadA-null cells. (D) 4-h developed wild-type cells. Note that after 4 h starvation, wild-type and mutant cells are indistinguishable. Bar, 10 μm.
Figure 6.
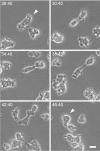
Axenically growing sadA-null cells. A cell going through an unsuccessful attempt to divide is marked by an arrow. The cell tried to pull apart, but finally “snapped” back together. Note that the whole process took more than 20 min, whereas a cell division in wild type is typically completed in 6–8 min. Note also that the cells display blebs and are neither attached nor spread (visible in the high refractive index of the light around the edges of the cells). For a better impression of the behavior of growing sadA-null cells, including the division of big cells into many daughters, and for a direct comparison with wild type, see the time-lapse videos (Videos 1 and 2, available online at
http://www.jcb.org/cgi/content/full/jcb.200206067/DC1
). Bar 10 μm.
Figure 7.
Growth and phagocytosis of sadA-null cells. (A) SadA-null cells grow faster in suspension. Two independent sadA-null strains (KO1 and KO2, open and closed inverted triangles, respectively) grow faster than wild type (open circles) and sadA–GFP-rescued mutant cells (closed circles). Doubling times were 10.1, 10.2, 12, and 13.5 h, respectively. After cells were established in suspension, growth was monitored for 3 d. (B) SadA-null cells have a strong phagocytosis defect. Cells were tested for the uptake rate of fluorescent latex beads over the course of 9 min. In sadA-null cells (closed circles), the uptake of beads was completely abolished. SadA–GFP-rescued mutant cells were sorted into high- (closed inverted triangles) and low-expressing (open inverted triangles) cells. In comparison to wild type (open circles), the low-expressing cells rescued the phagocytosis phenotype 64%, whereas the high-expressing cells showed an 83% rescue. For comparison, in myosin VII–null cells (closed squares), phagocytosis is reduced by ∼80% (see Discussion). (C and D) SadA is localized, but not enriched, in the phagocytic cup. SadA–GFP-rescued cells during an early (C) and later (D) stage of phagocytosis of heat-killed yeast particles (which show some autofluorescence).
Figure 7.
Growth and phagocytosis of sadA-null cells. (A) SadA-null cells grow faster in suspension. Two independent sadA-null strains (KO1 and KO2, open and closed inverted triangles, respectively) grow faster than wild type (open circles) and sadA–GFP-rescued mutant cells (closed circles). Doubling times were 10.1, 10.2, 12, and 13.5 h, respectively. After cells were established in suspension, growth was monitored for 3 d. (B) SadA-null cells have a strong phagocytosis defect. Cells were tested for the uptake rate of fluorescent latex beads over the course of 9 min. In sadA-null cells (closed circles), the uptake of beads was completely abolished. SadA–GFP-rescued mutant cells were sorted into high- (closed inverted triangles) and low-expressing (open inverted triangles) cells. In comparison to wild type (open circles), the low-expressing cells rescued the phagocytosis phenotype 64%, whereas the high-expressing cells showed an 83% rescue. For comparison, in myosin VII–null cells (closed squares), phagocytosis is reduced by ∼80% (see Discussion). (C and D) SadA is localized, but not enriched, in the phagocytic cup. SadA–GFP-rescued cells during an early (C) and later (D) stage of phagocytosis of heat-killed yeast particles (which show some autofluorescence).
Figure 8.
Vegetative sadA cells move faster than wild type. Following the paths of growing cells shows that sadA-null cells (A–F) move relatively fast (6.3 μm/min) (M), whereas wild-type cells (G–L) migrate with greater path persistence, at a slower rate (3.2 μm/min) (M). Six cells, shown in Videos 1 (sadA mutant) and 2 (wild type), available at
http://www.jcb.org/cgi/content/full/jcb.200206067/DC1
, were tracked.
Figure 9.
SadA is predominantly localized in the cortex. Fixed sadA–GFP-rescued mutant cells in transmitted and fluorescent light are shown. Note the different expression levels of sadA–GFP and that in some cells expression is very low or undetectable. Bar, 10 μm.
Figure 10.
Model summarizing the Dictyostelium proteins that are known to play a role in substrate adhesion. Myosin VII -, phg1-, and talin-null mutants all display an adhesion defect. Phg1 is a nine-transmembrane molecule involved in particle adhesion. While talin is known to link integrin to the cytoskeleton in higher organisms, it might link an adhesion receptor like sadA to the cytoskeleton in Dictyostelium. Myosin VII might be involved in the organization of molecules of the adhesion machinery. Activated rasG may regulate and activate a putative adhesion complex. SadA is the first putative adhesion receptor that is absolutely required for substrate adhesion in growth medium. It is possible that sadA, via its intracellular domain(s), is linked to the cytoskeleton. The extracellular EGF-like repeats (depicted in three black boxes) are prime candidates that may bind to external molecules, which may allow the cells to attach and spread.
Similar articles
- Uncovering a role for the tail of the Dictyostelium discoideum SadA protein in cell-substrate adhesion.
Kowal AS, Chisholm RL. Kowal AS, et al. Eukaryot Cell. 2011 May;10(5):662-71. doi: 10.1128/EC.00221-10. Epub 2011 Mar 25. Eukaryot Cell. 2011. PMID: 21441344 Free PMC article. - Requirement of a vasodilator-stimulated phosphoprotein family member for cell adhesion, the formation of filopodia, and chemotaxis in dictyostelium.
Han YH, Chung CY, Wessels D, Stephens S, Titus MA, Soll DR, Firtel RA. Han YH, et al. J Biol Chem. 2002 Dec 20;277(51):49877-87. doi: 10.1074/jbc.M209107200. Epub 2002 Oct 17. J Biol Chem. 2002. PMID: 12388544 - TM9/Phg1 and SadA proteins control surface expression and stability of SibA adhesion molecules in Dictyostelium.
Froquet R, le Coadic M, Perrin J, Cherix N, Cornillon S, Cosson P. Froquet R, et al. Mol Biol Cell. 2012 Feb;23(4):679-86. doi: 10.1091/mbc.E11-04-0338. Epub 2012 Jan 4. Mol Biol Cell. 2012. PMID: 22219373 Free PMC article. - Cell-cell signaling and adhesion in phagocytosis and early development of Dictyostelium.
Bracco E, Pergolizzi B, Peracino B, Ponte E, Balbo A, Mai A, Ceccarelli A, Bozzaro S. Bracco E, et al. Int J Dev Biol. 2000;44(6):733-42. Int J Dev Biol. 2000. PMID: 11061438 Review. - Cell adhesion in the life cycle of Dictyostelium.
Bozzaro S, Ponte E. Bozzaro S, et al. Experientia. 1995 Dec 18;51(12):1175-88. doi: 10.1007/BF01944735. Experientia. 1995. PMID: 8536805 Review.
Cited by
- An evolutionary and physiological perspective on cell-substrate adhesion machinery for cell migration.
Fierro Morales JC, Xue Q, Roh-Johnson M. Fierro Morales JC, et al. Front Cell Dev Biol. 2022 Aug 25;10:943606. doi: 10.3389/fcell.2022.943606. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36092727 Free PMC article. Review. - Cytoskeletal Mechanics Regulating Amoeboid Cell Locomotion.
Alvarez-González B, Meili R, Firtel R, Bastounis E, Del Álamo JC, Lasheras JC. Alvarez-González B, et al. Appl Mech Rev. 2014 Jun 5;66(5):0508041-05080414. doi: 10.1115/1.4026249. Appl Mech Rev. 2014. PMID: 25328163 Free PMC article. - Cooperative cell motility during tandem locomotion of amoeboid cells.
Bastounis E, Álvarez-González B, del Álamo JC, Lasheras JC, Firtel RA. Bastounis E, et al. Mol Biol Cell. 2016 Apr 15;27(8):1262-71. doi: 10.1091/mbc.E15-12-0836. Epub 2016 Feb 24. Mol Biol Cell. 2016. PMID: 26912787 Free PMC article. - Cell-to-cell signaling in cell populations with large cell size variability.
Hayashida Y, Oosawa C, Yasunaga T, Morimoto YV. Hayashida Y, et al. Biophys J. 2025 Mar 18;124(6):954-962. doi: 10.1016/j.bpj.2024.07.017. Epub 2024 Aug 12. Biophys J. 2025. PMID: 39137773 - A Rap/phosphatidylinositol 3-kinase pathway controls pseudopod formation [corrected].
Kortholt A, Bolourani P, Rehmann H, Keizer-Gunnink I, Weeks G, Wittinghofer A, Van Haastert PJ. Kortholt A, et al. Mol Biol Cell. 2010 Mar 15;21(6):936-45. doi: 10.1091/mbc.e09-03-0177. Epub 2010 Jan 20. Mol Biol Cell. 2010. PMID: 20089846 Free PMC article.
References
- Adachi, H., T. Hasebe, K. Yoshinaga, T. Ohta, and K. Sutoh. 1994. Isolation of Dictyostelium discoideum cytokinesis mutants by restriction enzyme-mediated integration of the blasticidin S resistance marker. Biochem. Biophys. Res. Commun. 205:1808–1814. - PubMed
- Brar, S., and C. Siu. 1993. Characterization of the cell adhesion molecule gp24 in Dictyostelium discoideum. Mediation of cell-cell adhesion via a Ca(2+)-dependent mechanism. J. Biol. Chem. 268:24902–24909. - PubMed
- Burridge, K., and L. Connell. 1983. Talin: a cytoskeletal component concentrated in adhesion plaques and other sites of actin-membrane interaction. Cell Motil. 3:405–417. - PubMed
- Burridge, K., and P. Mangeat. 1984. An interaction between vinculin and talin. Nature. 308:744–746. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials