Comparing the ex vivo fitness of CCR5-tropic human immunodeficiency virus type 1 isolates of subtypes B and C - PubMed (original) (raw)
Comparative Study
. 2003 Jan;77(2):1021-38.
doi: 10.1128/jvi.77.2.1021-1038.2003.
Awet Abraha, Kalonji R Collins, Andre J Marozsan, Heather Baird, Miguel E Quiñones-Mateu, Adam Penn-Nicholson, Michael Murray, Nathalie Richard, Michael Lobritz, Peter A Zimmerman, Tatsuyoshi Kawamura, Andrew Blauvelt, Eric J Arts
Affiliations
- PMID: 12502818
- PMCID: PMC140829
- DOI: 10.1128/jvi.77.2.1021-1038.2003
Comparative Study
Comparing the ex vivo fitness of CCR5-tropic human immunodeficiency virus type 1 isolates of subtypes B and C
Sarah C Ball et al. J Virol. 2003 Jan.
Abstract
Continual human immunodeficiency virus type 1 (HIV-1) evolution and expansion within the human population have led to unequal distribution of HIV-1 group M subtypes. In particular, recent outgrowth of subtype C in southern Africa, India, and China has fueled speculation that subtype C isolates may be more fit in vivo. In this study, nine subtype B and six subtype C HIV-1 isolates were added to peripheral blood mononuclear cell cultures for a complete pairwise competition experiment. All subtype C HIV-1 isolates were less fit than subtype B isolates (P < 0.0001), but intrasubtype variations in HIV-1 fitness were not significant. Increased fitness of subtype B over subtype C was also observed in primary CD4(+) T cells and macrophages from different human donors but not in skin-derived human Langerhans cells. Detailed analysis of the retroviral life cycle during several B and C virus competitions indicated that the efficiency of host cell entry may have a significant impact on relative fitness. Furthermore, phyletic analyses of fitness differences suggested that, for a recombined subtype B/C HIV-1 isolate, higher fitness mapped to the subtype B env gene rather than the subtype C gag and pol genes. These results suggest that subtype B and C HIV-1 may be transmitted with equal efficiency (Langerhans cell data) but that subtype C isolates may be less fit following initial infection (T-cell and macrophage data) and may lead to slower disease progression.
Figures
FIG. 1.
Measuring titers of HIV-1 stocks and replication kinetics. Four primary HIV-1 isolates, two subtype C and two subtype B strains (B2, B5, C2, and C5), were employed in monoinfection studies and are representative of the 15 isolates examined in this study. (A) Quantifying viral load in each stock by infection of PBMC with serially diluted virus (Reed-muench technique), by RT-PCR-amplifying HIV-1 RNA (3), and by measuring endogenous reverse transcriptase (RT) activity in virus stocks (77). Details of each assay are provided in the Materials and Methods section. (B) Coreceptor usage of each HIV-1 isolate. U87 cells expressing CD4 and CCR5 or CXCR4 were exposed to equal IU of each HIV-1 isolate. Virus production was measured by reverse transcriptase activity in the cell-free supernatant. All isolates except B4 were predicted to be R5/non-syncytium-inducing isolates, based on a more neutral V3 loop and neutral or negatively charged amino acids at position 306 and 322. (C) Virus production from PBMC monoinfected with 0.01 IU of B2, B5, C2, and C5 HIV-1 isolates. Reverse transcriptase activity in cell-free supernatant was measured during the 15-day monoinfection. (D) Virus production was measured by reverse transcriptase activity following a 15-day PBMC dual infection with B2, B5, C2, and C5. Two hundred and ten dual infections were analyzed with the same approach.
FIG. 2.
Strategy for HIV-1 competition experiments and heteroduplex tracking method for dual virus detection and quantification. (A) Virus was added alone or in pairs to phytohemagglutinin- and interleukin-2-treated PBMC at a multiplicity of infection (MOI) of 0.01. Cells were washed after 8 h to remove virus. Cells and virus supernatant was harvested at day 10 and lysed. (B) Extracted DNA or RNA from dual infections was PCR or RT-PCR amplified with conserved HIV-1 env primers. PCR-amplified env products were denatured, annealed to a radiolabeled env probe, and then run on an 8% nondenaturing polyacrylamide gel (C and D). Heteroduplexes generated from monoinfections (C) were then used to identify those isolates found in the heteroduplex tracking analysis (HTA) of each dual infection (D). Phosphor-imaging analysis of each heteroduplex was used to quantify the production of each virus in a dual infection. Multiple heteroduplexes can track to a single HIV-1 isolate because these isolates represent the propagated quasispecies and not a single clone.
FIG. 3.
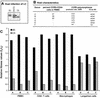
Pairwise competitions in PBMC to compare intrasubtype HIV-1 fitness differences. Nine subtype B and six subtype C HIV-1 isolates were employed in pairwise intrasubtype competitions. Fitness differences were plotted for the paired competitions between subtype B (panel A) and C (panel C) isolates. Fitness difference of each competition pair is presented as the fitness value of the isolate in the column divided by the fitness value of the isolate in the row. Average relative fitness values from these intrasubtype competitions in PBMC are provided in Table 1 and plotted with a phyletic analysis in Fig. 6.
FIG. 4.
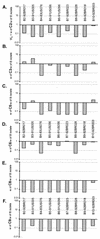
Comparing the relative fitness of subtype B and C HIV-1 isolates in direct competition. Subtype C isolates C2 (A), C3 (B), C5 (C), C6 (D), C8 (E), and C9 (F) were used to compete against each of the B isolates (B2, B3, B4, B5, B6, B8, B9, and B10) with equal multiplicities of infection for each isolate and PBMC cultures.
FIG. 5.
Comparing the relative fitness values of B5 and C5 HIV-1 isolates derived from competitions in CD4+ T cells, macrophages, and skin-derived Langerhans cells from different donors. (A) An example of B5 and C5 monoinfections and dual infection of skin-derived Langerhans cells (LC) from donor D. Skin explants were obtained from suction blister roofs isolated from the thighs of healthy donors (35) and exposed to a multiplicity of infection of 0.01 of the virus (250 IU added to approximately 25,000 Langerhans cells embedded within each skin explant). Langerhans cells were then allowed to emigrate from explants and were harvested on day 3 or 4 postinfection. DNA extracted from lysed cells was subjected to HIV-specific PCR amplifications and heteroduplex tracking analysis. Purification and subsequent HIV infections of macrophages and CD4+ T cells were done as described in Materials and Methods. Similar heteroduplex tracking analyses were performed on these samples. (B) Genetic characteristics and coreceptor expression on host PBMC cells. T cells and macrophages were purified from PBMC from three donors. Fluorescence-activated cell sorting analysis was performed on the PBMC populations prior to cell isolation to measure CCR5 and CD4 expression. PBMC labeled with fluorescein isothiocyanate-labeled anti-CD4 antibody and phycoerythrin-labeled anti-CCR5 antibodies were analyzed with the FACScan flow cytometer and Lysis II software (44). Percentages represent the fraction of CCR5-positive cells in the CD4-positive-gated lymphocyte population. Genetic polymorphisms at position −2459 in CCR5 promoter and the presence or absence of a 32-codon deletion (32 aa del) in the CCR5 open reading frame were also determined. wt, wild type; nd, not determined. (C) Relative fitness of the B5 and C5 HIV-1 isolates derived from dual infection of PBMC, CD4+ T cells, macrophages, and skin-derived Langerhans cells from different donors. Relative fitness value was measured as the proportion of each virus produced from the dual infection (_f_0) divided by the initial fraction of that virus added to the culture (_i_0) (62).
FIG. 6.
Time course competitions in PBMC with subtype B and C HIV-1 isolates and detection of HIV-1 products at different steps in the retroviral life cycle. (A) Schematic representation of pairwise dual infections with B2, B5, C2, and C5. PBMC were exposed to virus for 8 h, washed extensively, and reincubated at 37°C. Cells and supernatant were harvested at 8, 24, and 48 h and at 5 and 10 days postinfection. (B) Viral RNA or DNA extracted from infected cells was RT-PCR or PCR amplified with primer sets specific for early reverse transcripts, late reverse transcripts, integrated DNA, unspliced mRNA, and multiply spliced mRNA. Approximate positions of the primers are indicated. Radiolabeled primer pairs were used to detect and quantify products in both mono- and dual infections (C, D, and E), whereas an external-nested PCR amplification was necessary to generate sufficient product for heteroduplex tracking analysis (HTA) (Fig. 7). (C) Quantifying the level of HIV-1 integration during monoinfections with C5 and B5 HIV-1 isolates. HIV-1 DNA integrated within 4 kb of an Alu sequence in the human genome was PCR amplified with Alu and HIV-1 Alu-LTR primers (14). The primer pair LTR-1 and γ-32P-end-labeled AU3-1 was used for nested amplification of the previous PCR products. Time course samples for the C5 and B5 monoinfections were subjected to this external-nested PCR. To control for carryover of unintegrated HIV-1 DNA from the sample into the external and then nested PCR amplifications, 0.5 ml of the 240-h sample was added straight to the nested amplification (44). (D) Tenfold dilutions of linearized HXB2 plasmid (108 to 102) were PCR amplified with the nested primer pair as an amplification control and to estimate copy number for each specific product. Similar amplifications were performed on the different RNA and DNA HIV-1 products (A and B). However, in vitro-transcribed HIV-1 RNA was quantified, diluted, and used as template for control RT-PCR amplifications (data not shown). Copy number of sample RNA/DNA was derived from amplifications of known copies of control template. Copy number of control DNA and resulting PCR-amplified early, late, or integrated DNA product was plotted to obtain an equation of the line. All power regressions had _R_2 values of greater than 0.95 with a template copy range of 102 to 106. (E) Approximate copy numbers of early and late reverse-transcribed DNA, integrated DNA, unspliced mRNA, and multiply spliced mRNA from the C5 monoinfection were plotted against time. Similar results were observed in the B5 monoinfection as well as in the B5-versus-C5 dual infection (data not shown). (-), minus strand; (-)ss, minus-strand strong stop.
FIG. 6.
Time course competitions in PBMC with subtype B and C HIV-1 isolates and detection of HIV-1 products at different steps in the retroviral life cycle. (A) Schematic representation of pairwise dual infections with B2, B5, C2, and C5. PBMC were exposed to virus for 8 h, washed extensively, and reincubated at 37°C. Cells and supernatant were harvested at 8, 24, and 48 h and at 5 and 10 days postinfection. (B) Viral RNA or DNA extracted from infected cells was RT-PCR or PCR amplified with primer sets specific for early reverse transcripts, late reverse transcripts, integrated DNA, unspliced mRNA, and multiply spliced mRNA. Approximate positions of the primers are indicated. Radiolabeled primer pairs were used to detect and quantify products in both mono- and dual infections (C, D, and E), whereas an external-nested PCR amplification was necessary to generate sufficient product for heteroduplex tracking analysis (HTA) (Fig. 7). (C) Quantifying the level of HIV-1 integration during monoinfections with C5 and B5 HIV-1 isolates. HIV-1 DNA integrated within 4 kb of an Alu sequence in the human genome was PCR amplified with Alu and HIV-1 Alu-LTR primers (14). The primer pair LTR-1 and γ-32P-end-labeled AU3-1 was used for nested amplification of the previous PCR products. Time course samples for the C5 and B5 monoinfections were subjected to this external-nested PCR. To control for carryover of unintegrated HIV-1 DNA from the sample into the external and then nested PCR amplifications, 0.5 ml of the 240-h sample was added straight to the nested amplification (44). (D) Tenfold dilutions of linearized HXB2 plasmid (108 to 102) were PCR amplified with the nested primer pair as an amplification control and to estimate copy number for each specific product. Similar amplifications were performed on the different RNA and DNA HIV-1 products (A and B). However, in vitro-transcribed HIV-1 RNA was quantified, diluted, and used as template for control RT-PCR amplifications (data not shown). Copy number of sample RNA/DNA was derived from amplifications of known copies of control template. Copy number of control DNA and resulting PCR-amplified early, late, or integrated DNA product was plotted to obtain an equation of the line. All power regressions had _R_2 values of greater than 0.95 with a template copy range of 102 to 106. (E) Approximate copy numbers of early and late reverse-transcribed DNA, integrated DNA, unspliced mRNA, and multiply spliced mRNA from the C5 monoinfection were plotted against time. Similar results were observed in the B5 monoinfection as well as in the B5-versus-C5 dual infection (data not shown). (-), minus strand; (-)ss, minus-strand strong stop.
FIG. 7.
Measuring isolate-specific products that represent different steps in the retroviral life cycle during a dual-infection/competition experiment. Relative production of two HIV-1 isolates was measured during different steps in the HIV-1 life cycle, i.e., early and late reverse transcription, integration, and unspliced and multiply spliced mRNA transcription. Panel A provides an example of one heteroduplex tracking analysis performed on the late product of HIV-1 reverse transcription that was PCR amplified at 120 h postinfection. (-), minus strand. The products in the lanes identified with bold and italic labels were quantified and plotted in panels A, B, and C. These panels show the results of three time course dual infections involving B5 versus C5, B2 versus C5, and B5 versus B2, respectively.
FIG. 8.
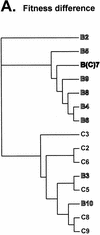
Analysis of phenotypic and genotypic relationships of HIV-1 isolates with the neighbor-joining method. Phyletic neighbor-joining trees were constructed from matrices of pairwise log10 fitness differences as well as from nucleotide distances in the env and gag genes. (A) The neighbor.exe program in the PHYLIP package was used to draw a phyletic tree based on the log10 values of the fitness difference in the pairwise dual-infection/competition experiments. Equal relative fitness values result in a fitness difference of 1. Thus, log10 values of w M/w L provide a unidirectional measure of fitness difference for comparison to genotypic distances. (B and C) Phylogenetic analyses of the gag and env sequences, respectively, of 15 subtype B and C isolates and of several reference HIV-1 isolates (see Materials and Methods for accession numbers). A 400-bp region of the env gene C2-C3 region (B) and a 500-bp region of the gag gene (C) were used to construct phylogenetic trees by the neighbor-joining method. env and gag subtypes are indicated. Bootstrap resampling values of 70 to 90% and >90% are indicated by ** and *, respectively. The branch lengths are drawn to scale. The scale bar represents 0.1 substitution per nucleotide.
FIG. 8.
Analysis of phenotypic and genotypic relationships of HIV-1 isolates with the neighbor-joining method. Phyletic neighbor-joining trees were constructed from matrices of pairwise log10 fitness differences as well as from nucleotide distances in the env and gag genes. (A) The neighbor.exe program in the PHYLIP package was used to draw a phyletic tree based on the log10 values of the fitness difference in the pairwise dual-infection/competition experiments. Equal relative fitness values result in a fitness difference of 1. Thus, log10 values of w M/w L provide a unidirectional measure of fitness difference for comparison to genotypic distances. (B and C) Phylogenetic analyses of the gag and env sequences, respectively, of 15 subtype B and C isolates and of several reference HIV-1 isolates (see Materials and Methods for accession numbers). A 400-bp region of the env gene C2-C3 region (B) and a 500-bp region of the gag gene (C) were used to construct phylogenetic trees by the neighbor-joining method. env and gag subtypes are indicated. Bootstrap resampling values of 70 to 90% and >90% are indicated by ** and *, respectively. The branch lengths are drawn to scale. The scale bar represents 0.1 substitution per nucleotide.
Similar articles
- CCR5- and CXCR4-tropic subtype C human immunodeficiency virus type 1 isolates have a lower level of pathogenic fitness than other dominant group M subtypes: implications for the epidemic.
Abraha A, Nankya IL, Gibson R, Demers K, Tebit DM, Johnston E, Katzenstein D, Siddiqui A, Herrera C, Fischetti L, Shattock RJ, Arts EJ. Abraha A, et al. J Virol. 2009 Jun;83(11):5592-605. doi: 10.1128/JVI.02051-08. Epub 2009 Mar 18. J Virol. 2009. PMID: 19297481 Free PMC article. - Differences in the fitness of two diverse wild-type human immunodeficiency virus type 1 isolates are related to the efficiency of cell binding and entry.
Marozsan AJ, Moore DM, Lobritz MA, Fraundorf E, Abraha A, Reeves JD, Arts EJ. Marozsan AJ, et al. J Virol. 2005 Jun;79(11):7121-34. doi: 10.1128/JVI.79.11.7121-7134.2005. J Virol. 2005. PMID: 15890952 Free PMC article. - Genetic analysis of HIV-1 isolates from Brazil reveals presence of two distinct genetic subtypes.
Louwagie J, Delwart EL, Mullins JI, McCutchan FE, Eddy G, Burke DS. Louwagie J, et al. AIDS Res Hum Retroviruses. 1994 May;10(5):561-7. doi: 10.1089/aid.1994.10.561. AIDS Res Hum Retroviruses. 1994. PMID: 7917518 - The predominance of Human Immunodeficiency Virus type 1 (HIV-1) circulating recombinant form 02 (CRF02_AG) in West Central Africa may be related to its replicative fitness.
Njai HF, Gali Y, Vanham G, Clybergh C, Jennes W, Vidal N, Butel C, Mpoudi-Ngolle E, Peeters M, Ariën KK. Njai HF, et al. Retrovirology. 2006 Jul 3;3:40. doi: 10.1186/1742-4690-3-40. Retrovirology. 2006. PMID: 16817969 Free PMC article. - High replication fitness and transmission efficiency of HIV-1 subtype C from India: Implications for subtype C predominance.
Rodriguez MA, Ding M, Ratner D, Chen Y, Tripathy SP, Kulkarni SS, Chatterjee R, Tarwater PM, Gupta P. Rodriguez MA, et al. Virology. 2009 Mar 15;385(2):416-24. doi: 10.1016/j.virol.2008.12.025. Epub 2009 Jan 20. Virology. 2009. PMID: 19157481
Cited by
- CD4+ T Cell-Mimicking Nanoparticles Broadly Neutralize HIV-1 and Suppress Viral Replication through Autophagy.
Zhang G, Campbell GR, Zhang Q, Maule E, Hanna J, Gao W, Zhang L, Spector SA. Zhang G, et al. mBio. 2020 Sep 15;11(5):e00903-20. doi: 10.1128/mBio.00903-20. mBio. 2020. PMID: 32934078 Free PMC article. - Molecular and phylogeographic analysis of human immuno-deficiency virus type 1 strains infecting treatment-naive patients from Kigali, Rwanda.
Rusine J, Jurriaans S, van de Wijgert J, Cornelissen M, Kateera B, Boer K, Karita E, Mukabayire O, de Jong M, Ondoa P. Rusine J, et al. PLoS One. 2012;7(8):e42557. doi: 10.1371/journal.pone.0042557. Epub 2012 Aug 14. PLoS One. 2012. PMID: 22905148 Free PMC article. - Human Three-Dimensional Models for Studying Skin Pathogens.
Boero E, Mnich ME, Manetti AGO, Soldaini E, Grimaldi L, Bagnoli F. Boero E, et al. Curr Top Microbiol Immunol. 2021;430:3-27. doi: 10.1007/82_2020_219. Curr Top Microbiol Immunol. 2021. PMID: 32601967 Review. - Characterization of a subtype D human immunodeficiency virus type 1 isolate that was obtained from an untreated individual and that is highly resistant to nonnucleoside reverse transcriptase inhibitors.
Gao Y, Paxinos E, Galovich J, Troyer R, Baird H, Abreha M, Kityo C, Mugyenyi P, Petropoulos C, Arts EJ. Gao Y, et al. J Virol. 2004 May;78(10):5390-401. doi: 10.1128/jvi.78.10.5390-5401.2004. J Virol. 2004. PMID: 15113918 Free PMC article. - The replicative fitness of primary human immunodeficiency virus type 1 (HIV-1) group M, HIV-1 group O, and HIV-2 isolates.
Ariën KK, Abraha A, Quiñones-Mateu ME, Kestens L, Vanham G, Arts EJ. Ariën KK, et al. J Virol. 2005 Jul;79(14):8979-90. doi: 10.1128/JVI.79.14.8979-8990.2005. J Virol. 2005. PMID: 15994792 Free PMC article.
References
- Alaeus, A., K. Lidman, A. Bjorkman, J. Giesecke, and J. Albert. 1999. Similar rate of disease progression among individuals infected with HIV-1 genetic subtypes A-D. AIDS 13:901-907. - PubMed
- Reference deleted.
- Arts, E. J., J. Mak, L. Kleiman, and M. A. Wainberg. 1994. DNA found in human immunodeficiency virus type 1 particles may not be required for infectivity. J. Gen. Virol. 75:1605-1613. - PubMed
- Asjo, B., L. Morfeldt-Manson, J. Albert, G. Biberfeld, A. Karlsson, K. Lidman, and E. M. Fenyo. 1986. Replicative capacity of human immunodeficiency virus from patients with varying severity of HIV infection. Lancet 2:660-662. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R21 AI049170/AI/NIAID NIH HHS/United States
- AI49170/AI/NIAID NIH HHS/United States
- R56 AI049170/AI/NIAID NIH HHS/United States
- AI36219/AI/NIAID NIH HHS/United States
- N01-HD-0-3310-502-02/HD/NICHD NIH HHS/United States
- R01 AI049170/AI/NIAID NIH HHS/United States
- K01 HL067610/HL/NHLBI NIH HHS/United States
- HL67610-02/HL/NHLBI NIH HHS/United States
- T32 GM007250/GM/NIGMS NIH HHS/United States
- P30 AI036219/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials