Antiviral effect and virus-host interactions in response to alpha interferon, gamma interferon, poly(i)-poly(c), tumor necrosis factor alpha, and ribavirin in hepatitis C virus subgenomic replicons - PubMed (original) (raw)
Antiviral effect and virus-host interactions in response to alpha interferon, gamma interferon, poly(i)-poly(c), tumor necrosis factor alpha, and ribavirin in hepatitis C virus subgenomic replicons
Robert E Lanford et al. J Virol. 2003 Jan.
Abstract
The recently developed hepatitis C virus (HCV) subgenomic replicon system was utilized to evaluate the efficacy of several known antiviral agents. Cell lines that persistently maintained a genotype 1b replicon were selected. The replicon resident in each cell line had acquired adaptive mutations in the NS5A region that increased colony-forming efficiency, and some replicons had acquired NS3 mutations that alone did not enhance colony-forming efficiency but were synergistic with NS5A mutations. A replicon constructed from the infectious clone of the HCV-1 strain (genotype 1a) was not capable of inducing colony formation even after the introduction of adaptive mutations identified in the genotype 1b replicon. Alpha interferon (IFN-alpha), IFN-gamma, and ribavirin exhibited antiviral activity, while double-stranded RNA (dsRNA) and tumor necrosis factor alpha did not. Analysis of transcript levels for a series of genes stimulated by IFN (ISGs) or dsRNA following treatment with IFN-alpha, IFN-gamma, and dsRNA revealed that both IFNs increased ISG transcript levels, but that some aspect of the dsRNA response pathway was defective in Huh7 cells and replicon cell lines in comparison to primary chimpanzee and tamarin hepatocytes. The colony-forming efficiency of the replicon was reduced or eliminated following replication in the presence of ribavirin, implicating the induction of error-prone replication. The potential role of error-prone replication in the synergy observed between IFN-alpha and ribavirin in attaining sustained viral clearance is discussed. These studies reveal characteristics of Huh7 cells that may contribute to their unique capacity to support HCV RNA synthesis and demonstrate the utility of the replicon system for mechanistic studies on antiviral agents.
Figures
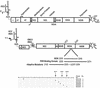
FIG. 1.
Schematic of HCV genome and replicon design. The HCV genome is depicted with the 5′ NCR containing an IRES and the 3′ NCR including a variable region (Var), a polyuridine or polypyrimidine stretch (U/PP), and a 98-nucleotide (nt) conserved region (CR). The depiction of the HCV IRES was adapted from the structural study of Honda et al. (28). The open reading frame of the polyprotein is depicted as a rectangle with demarcation of the individual viral protein domains, and the positions of some of the viral functions are depicted above. c, capsid protein; e1 and e2, envelope proteins E1 and E2. The structure of the bicistronic replicon is illustrated below the HCV genome, with the HCV IRES and EMCV IRES regulating translation of the neomycin phosphotransferase (NEO) gene and the nonstructural proteins of HCV, respectively. A T7 promoter is fused to the HCV 5′ terminus for the production of synthetic RNA. A domain of the NS5A protein is expanded to illustrate the ISDR, PKR binding domain, and the region of the most common adaptive replicon mutations, including the deletion from amino acids 2207 to 2254 observed by Blight et al. (8). The amino acid changes for the adaptive mutations detected in the resident replicons isolated from 10 independent G418-resistant colonies are indicated at the bottom of the figure, and the sites of NS5A hyperphosphorylation (amino acids 2197, 2201, and 2204) are indicated by arrows. WT, wild type.
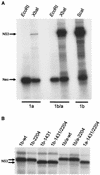
FIG. 2.
In vitro translation of bicistronic HCV replicons. (A) Rep1aNeo (1a), Rep1b/a (1b/a), and Rep1b (1b) synthetic RNAs were produced by in vitro transcription reactions utilizing vectors linearized at _Eco_RI, a site downstream of the neomycin phosphotransferase gene (Fig. 1), or at _Xba_I or _Sca_I to produce full-length replicon RNA. RNAs were in vitro translated using rabbit reticulocyte lysates in the presence of [35S]methionine, and the products were analyzed by SDS-PAGE. Rep1a produced reduced levels of NS3 compared to both Rep1b and Rep1b/a, while translation of the Neo protein was similar for all constructs. (B) The impact of the NS3 D1431Y mutation on the migration of NS3 by SDS-PAGE was examined following in vitro translation. All replicon RNAs containing the D1431Y mutation exhibited an increased mobility for NS3 regardless of whether the mutation was in the presence of the NS5A S2204I mutation or in the Rep1b (1b) or Rep1a (1a) background. wt, wild type.
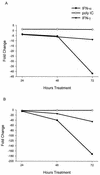
FIG. 3.
Kinetics of HCV replicon RNA decline following treatment with IFN-α, IFN-γ, and poly(I)-poly(C). Clone 45 cells were treated with IFN-α or IFN-γ at 100 or 1,000 U/ml and with poly(I)-poly(C) at 100 or 1,000 μg/ml and harvested at 24, 48, and 72 h. Results from 100 (A) and 1,000 (B) U/ml or μg/ml. Replicon RNA levels were quantified by TaqMan RT-PCR and expressed as the fold change from the levels of untreated cultures harvested at the same time points. All values are averages from duplicate cultures.
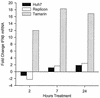
FIG. 4.
IFN-β transcription in response to poly(I)-poly(C). Huh7, clone 45, and primary tamarin hepatocyte cultures were harvested after 2, 7, and 24 h of treatment with 100 μg of poly(I)-poly(C) per ml. The levels of IFN-β transcripts were quantified in total cell RNA by TaqMan RT-PCR and were expressed as fold change in comparison to the levels of untreated cells. All values are averages of duplicate cultures.
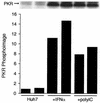
FIG. 5.
PKR activation by poly(I)-poly(C) in Huh7 cells. Huh7 cells were treated with 100 μg of poly(I)-poly(C) per ml for 2 h or with 1,000 U of IFN-α per ml for 16 h prior to harvest. PKR was immunoprecipitated from cell lysates, and kinase reactions were conducted with [γ-32P]ATP with PKR still bound to the antibody beads. Phosphorylated PKR was analyzed by SDS-PAGE and autoradiography (top) or by phosphorimage analysis (bottom). Phosphorimage values are expressed as arbitrary units and represent the volume in individual PKR bands shown in the blot.
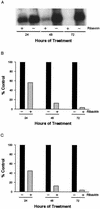
FIG. 6.
Antiviral effect of ribavirin on HCV replicon RNA. Clone 24 cells were cultivated with (+) or without (−) 400 μM ribavirin for 24, 48, or 72 h. Replicon RNA was analyzed by agarose gel electrophoresis, Northern blot hybridization, and autoradiography (A). RNA in the same blot was quantified by phosphorimage analysis (B), or the replicon RNA was quantified by TaqMan RT-PCR (C). Values in panels B and C are expressed as percentages of untreated, control cultures at each time point.
Similar articles
- Replication studies using genotype 1a subgenomic hepatitis C virus replicons.
Gu B, Gates AT, Isken O, Behrens SE, Sarisky RT. Gu B, et al. J Virol. 2003 May;77(9):5352-9. doi: 10.1128/jvi.77.9.5352-5359.2003. J Virol. 2003. PMID: 12692237 Free PMC article. - Inhibition of subgenomic hepatitis C virus RNA in Huh-7 cells: ribavirin induces mutagenesis in HCV RNA.
Kanda T, Yokosuka O, Imazeki F, Tanaka M, Shino Y, Shimada H, Tomonaga T, Nomura F, Nagao K, Ochiai T, Saisho H. Kanda T, et al. J Viral Hepat. 2004 Nov;11(6):479-87. doi: 10.1111/j.1365-2893.2004.00531.x. J Viral Hepat. 2004. PMID: 15500548 - Identification of a ribavirin-resistant NS5B mutation of hepatitis C virus during ribavirin monotherapy.
Young KC, Lindsay KL, Lee KJ, Liu WC, He JW, Milstein SL, Lai MM. Young KC, et al. Hepatology. 2003 Oct;38(4):869-78. doi: 10.1053/jhep.2003.50445. Hepatology. 2003. PMID: 14512874 Clinical Trial. - Understanding the molecular mechanism(s) of hepatitis C virus (HCV) induced interferon resistance.
Qashqari H, Al-Mars A, Chaudhary A, Abuzenadah A, Damanhouri G, Alqahtani M, Mahmoud M, El Sayed Zaki M, Fatima K, Qadri I. Qashqari H, et al. Infect Genet Evol. 2013 Oct;19:113-9. doi: 10.1016/j.meegid.2013.06.025. Epub 2013 Jul 5. Infect Genet Evol. 2013. PMID: 23831932 Review. - [Interferon resistance and ISDR (interferon sensitivity determining region)].
Amemiya F, Maekawa S, Enomoto N. Amemiya F, et al. Nihon Rinsho. 2006 Jul;64(7):1249-53. Nihon Rinsho. 2006. PMID: 16838640 Review. Japanese.
Cited by
- Modulation of host metabolism as a target of new antivirals.
Ikeda M, Kato N. Ikeda M, et al. Adv Drug Deliv Rev. 2007 Oct 10;59(12):1277-89. doi: 10.1016/j.addr.2007.03.021. Epub 2007 Aug 11. Adv Drug Deliv Rev. 2007. PMID: 17897752 Free PMC article. Review. - Negative regulation of intracellular hepatitis C virus replication by interferon regulatory factor 3.
Yamashiro T, Sakamoto N, Kurosaki M, Kanazawa N, Tanabe Y, Nakagawa M, Chen CH, Itsui Y, Koyama T, Takeda Y, Maekawa S, Enomoto N, Sakugawa H, Watanabe M. Yamashiro T, et al. J Gastroenterol. 2006 Aug;41(8):750-7. doi: 10.1007/s00535-006-1842-x. J Gastroenterol. 2006. PMID: 16988763 - Essential Role of Interferon Response in Containing Human Pathogenic Bourbon Virus.
Fuchs J, Straub T, Seidl M, Kochs G. Fuchs J, et al. Emerg Infect Dis. 2019 Jul;25(7):1304-1313. doi: 10.3201/eid2507.181062. Emerg Infect Dis. 2019. PMID: 31211667 Free PMC article. - New strategies for the treatment of hepatitis C virus infection and implications of resistance to new direct-acting antiviral agents.
Quer J, Buti M, Cubero M, Guardia J, Esteban R, Esteban JI. Quer J, et al. Infect Drug Resist. 2010;3:133-45. doi: 10.2147/IDR.S7136. Epub 2010 Nov 23. Infect Drug Resist. 2010. PMID: 21694902 Free PMC article. - Therapeutic implications of hepatitis C virus resistance to antiviral drugs.
Pawlotsky JM. Pawlotsky JM. Therap Adv Gastroenterol. 2009 Jul;2(4):205-19. doi: 10.1177/1756283X09336045. Therap Adv Gastroenterol. 2009. PMID: 21180544 Free PMC article.
References
- Alter, H. J., and M. Houghton. 2000. Hepatitis C virus and eliminating post-transfusion hepatitis. Nat. Med. 6:1082-1086. - PubMed
- Alter, H. J., and L. B. Seeff. 2000. Recovery, persistence, and sequelae in hepatitis C virus infection: a perspective on long-term outcome. Semin. Liver Dis. 20:17-35. - PubMed
- Alter, M. J., D. Kruszon-Moran, O. V. Nainan, G. M. McQuillan, F. X. Gao, L. A. Moyer, R. A. Kaslow, and H. S. Margolis. 1999. The prevalence of hepatitis C virus infection in the United States, 1988 through 1994. N. Engl. J. Med. 341:556-562. - PubMed
- Bandyopadhyay, S. K., G. T. Leonard, Jr., T. Bandyopadhyay, G. R. Stark, and G. C. Sen. 1995. Transcriptional induction by double-stranded RNA is mediated by interferon-stimulated response elements without activation of interferon-stimulated gene factor 3. J. Biol. Chem. 270:19624-19629. - PubMed
- Bartenschlager, R., and V. Lohmann. 2000. Replication of hepatitis C virus. J. Gen. Virol. 81:1631-1648. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U19 AI040035/AI/NIAID NIH HHS/United States
- P51 RR13986/RR/NCRR NIH HHS/United States
- U19 AI40035/AI/NIAID NIH HHS/United States
- R01 AI049574/AI/NIAID NIH HHS/United States
- R01 AI49574/AI/NIAID NIH HHS/United States
- P51 RR013986/RR/NCRR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources