Binding dynamics of structural nucleoporins govern nuclear pore complex permeability and may mediate channel gating - PubMed (original) (raw)
Binding dynamics of structural nucleoporins govern nuclear pore complex permeability and may mediate channel gating
Nataliya Shulga et al. Mol Cell Biol. 2003 Jan.
Abstract
The nuclear pore complex (NPC) is a permeable sieve that can dilate to facilitate the bidirectional translocation of a wide size range of receptor-cargo complexes. The binding of receptors to FG nucleoporin docking sites triggers channel gating by an unknown mechanism. Previously, we used deoxyglucose and chilling treatments to implicate Nup170p and Nup188p in the control of NPC sieving in Saccharomyces cerevisiae. Here, we report that aliphatic alcohols increase the permeability of wild-type and nup170Delta NPCs. In conjunction with increases in permeability, aliphatic alcohols, deoxyglucose, and chilling trigger the reversible dissociation of several nucleoporins from nup170Delta NPCs. These results are consistent with the hypothesis that NPC gating occurs when molecular latches composed of FG repeats and structural nucleoporins dissociate.
Figures
FIG. 1.
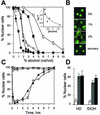
Aliphatic alcohols promote equilibration of cNLS-GFP. (A) Dose-dependent capacity of various alcohols to induce the equilibration of cNLS-GFP across wt nuclear envelopes. ○, methanol; ◊, ethanol; ▪, isopropanol; ▴, HD; •, _n_-butanol. Cells were exposed to the alcohols mixed with SC-Glu, and the percentage of cells exhibiting predominately nuclear localization of cNLS-GFP (% Nuclear cells) after 10 min was plotted as a function of alcohol concentration (vol/vol). The inset shows the relationship between the relative hydrophobicities (log P) (29) of five alcohols and the molar concentrations at which each induced 50% delocalization of cNLS-GFP in 10 min. Data for cyclohexanol (▵) are included instead of data for HD, for which a partition coefficient is not available. (B) Confocal images of wt cells treated with 0, 1, and 2% HD for 10 min and of cells after a 30-min recovery after 10 min in 5% alcohol. (C) Time course of cNLS-GFP localization in wt cells incubated in either 5% ethanol (▴) or 2% HD (⋄). Also shown are data for cells expressing GAL1::SSA1 in 5% ethanol (▵) or 2% HD (⋄). (D) Adaptive response of wt cells continuously incubated for 6 h in either 2% HD or 5% ethanol. Untreated control cells (white bars), HD-pretreated cells (grey bars), and ethanol-pretreated cells (black bars) were subsequently challenged with either 2% HD or 5% ethanol (EtOH) for 10 min before cNLS-GFP localization was scored.
FIG. 2.
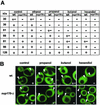
Alcohol-induced increases in the sieving diameters of wt and _nup170_Δ NPCs. (A) Cells expressing NES-GFP reporters of various sizes were incubated for 10 min in the presence of 5% concentrations of ethanol, 2-propanol, or HD or 2% butanol. Minus signs indicate that the reporter was not excluded from the large majority of nuclei, plus signs indicate that the reporter was excluded from nuclei, and +− indicates partial equilibration. (B) Confocal images of wt and _nup170_Δ cells expressing the 66-kDa NES-GFP reporter and treated for 10 min with each of the indicated alcohols at the concentrations stated above. Arrowheads indicate location of nuclei. Scoring of cells is indicated in the lower right corner of each panel. The large black vacuolar compartments serve as a control for complete exclusion of the GFP reporter.
FIG. 3.
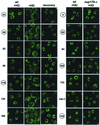
Reversible dissociation of nups in _nup170_Δ cells is induced by HD. Shown are confocal images revealing the localization of 14 different Nup-EYFP and Nup-GFP reporters in wt and _nup170_Δ cells after 10 min in the presence of 5% HD or after recovery from the alcohol (see Material and Methods). Nups are indicated by the numbers associated with their gene designations (e.g., 53 indicates Nup53p-EYFP), and the numbers for Nups containing FG repeats are circled.
FIG. 4.
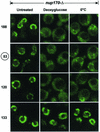
Dissociation of nups in _nup170_Δ cells induced by deoxyglucose and chilling. Shown are confocal images revealing the localization of four Nup-EYFP reporters in _nup170_Δ cells in SC-Glu medium before (untreated) and after treatment with 20 mM deoxyglucose for 30 min at 30°C (Deoxyglucose) or chilled on ice for 30 min (0°C). The nups are designated as described in the legend to Fig. 3.
Similar articles
- Natively unfolded nucleoporins gate protein diffusion across the nuclear pore complex.
Patel SS, Belmont BJ, Sante JM, Rexach MF. Patel SS, et al. Cell. 2007 Apr 6;129(1):83-96. doi: 10.1016/j.cell.2007.01.044. Cell. 2007. PMID: 17418788 - Nuclear mRNA export requires specific FG nucleoporins for translocation through the nuclear pore complex.
Terry LJ, Wente SR. Terry LJ, et al. J Cell Biol. 2007 Sep 24;178(7):1121-32. doi: 10.1083/jcb.200704174. Epub 2007 Sep 17. J Cell Biol. 2007. PMID: 17875746 Free PMC article. - The permeability of reconstituted nuclear pores provides direct evidence for the selective phase model.
Hülsmann BB, Labokha AA, Görlich D. Hülsmann BB, et al. Cell. 2012 Aug 17;150(4):738-51. doi: 10.1016/j.cell.2012.07.019. Cell. 2012. PMID: 22901806 - Structural dynamics of the nuclear pore complex.
Sakiyama Y, Panatala R, Lim RYH. Sakiyama Y, et al. Semin Cell Dev Biol. 2017 Aug;68:27-33. doi: 10.1016/j.semcdb.2017.05.021. Epub 2017 Jun 1. Semin Cell Dev Biol. 2017. PMID: 28579449 Review. - Lighting up the nuclear pore complex.
Kahms M, Hüve J, Wesselmann R, Farr JC, Baumgärtel V, Peters R. Kahms M, et al. Eur J Cell Biol. 2011 Sep;90(9):751-8. doi: 10.1016/j.ejcb.2011.04.004. Epub 2011 May 31. Eur J Cell Biol. 2011. PMID: 21632146 Review.
Cited by
- Nup98 FG domains from diverse species spontaneously phase-separate into particles with nuclear pore-like permselectivity.
Schmidt HB, Görlich D. Schmidt HB, et al. Elife. 2015 Jan 6;4:e04251. doi: 10.7554/eLife.04251. Elife. 2015. PMID: 25562883 Free PMC article. - The chaperone DNAJB6 surveils FG-nucleoporins and is required for interphase nuclear pore complex biogenesis.
Kuiper EFE, Gallardo P, Bergsma T, Mari M, Kolbe Musskopf M, Kuipers J, Giepmans BNG, Steen A, Kampinga HH, Veenhoff LM, Bergink S. Kuiper EFE, et al. Nat Cell Biol. 2022 Nov;24(11):1584-1594. doi: 10.1038/s41556-022-01010-x. Epub 2022 Oct 27. Nat Cell Biol. 2022. PMID: 36302971 - Biology and biophysics of the nuclear pore complex and its components.
Lim RY, Ullman KS, Fahrenkrog B. Lim RY, et al. Int Rev Cell Mol Biol. 2008;267:299-342. doi: 10.1016/S1937-6448(08)00632-1. Int Rev Cell Mol Biol. 2008. PMID: 18544502 Free PMC article. Review. - Unveiling the complexity: assessing models describing the structure and function of the nuclear pore complex.
Rush C, Jiang Z, Tingey M, Feng F, Yang W. Rush C, et al. Front Cell Dev Biol. 2023 Oct 9;11:1245939. doi: 10.3389/fcell.2023.1245939. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 37876551 Free PMC article. Review. - Size-dependent leak of soluble and membrane proteins through the yeast nuclear pore complex.
Popken P, Ghavami A, Onck PR, Poolman B, Veenhoff LM. Popken P, et al. Mol Biol Cell. 2015 Apr 1;26(7):1386-94. doi: 10.1091/mbc.E14-07-1175. Epub 2015 Jan 28. Mol Biol Cell. 2015. PMID: 25631821 Free PMC article.
References
- Aitchison, J. D., M. P. Rout, M. Marelli, G. Blobel, and R. W. Wozniak. 1995. Two novel related yeast nucleoporins Nup170p and Nup157p: complementation with the vertebrate homologue Nup155p and functional interactions with the yeast nuclear pore-membrane protein Pom152p. J. Cell Biol. 131:1133-1148. - PMC - PubMed
- Akey, C. W. 1995. Structural plasticity of the nuclear pore complex. J. Mol. Biol. 248:273-293. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases