Evaluation of PCR amplification bias by terminal restriction fragment length polymorphism analysis of small-subunit rRNA and mcrA genes by using defined template mixtures of methanogenic pure cultures and soil DNA extracts - PubMed (original) (raw)
Evaluation of PCR amplification bias by terminal restriction fragment length polymorphism analysis of small-subunit rRNA and mcrA genes by using defined template mixtures of methanogenic pure cultures and soil DNA extracts
Tillmann Lueders et al. Appl Environ Microbiol. 2003 Jan.
Abstract
Terminal restriction fragment length polymorphism (T-RFLP) analysis is a widely used method for profiling microbial community structure in different habitats by targeting small-subunit (SSU) rRNA and also functional marker genes. It is not known, however, whether relative gene frequencies of individual community members are adequately represented in post-PCR amplicon frequencies as shown by T-RFLP. In this study, precisely defined artificial template mixtures containing genomic DNA of four different methanogens in various ratios were prepared for subsequent T-RFLP analysis. PCR amplicons were generated from defined mixtures targeting not only the SSU rRNA but also the methyl-coenzyme M reductase (mcrA/mrtA) genes of methanogens. Relative amplicon frequencies of microorganisms were quantified by comparing fluorescence intensities of characteristic terminal restriction fragments. SSU ribosomal DNA (rDNA) template ratios in defined template mixtures of the four-membered community were recovered absolutely by PCR-T-RFLP analysis, which demonstrates that the T-RFLP analysis evaluated can give a quantitative view of the template pool. SSU rDNA-targeted T-RFLP analysis of a natural community was found to be highly reproducible, independent of PCR annealing temperature, and unaffected by increasing PCR cycle numbers. Ratios of mcrA-targeted T-RFLP analysis were biased, most likely by PCR selection due to the degeneracy of the primers used. Consequently, for microbial community analyses, each primer system used should be evaluated carefully for possible PCR bias. In fact, such bias can be detected by using T-RFLP analysis as a tool for the precise quantification of the PCR product pool.
Figures
FIG. 1.
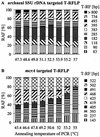
Effect of various PCR annealing temperatures on the SSU rDNA-targeted (A) and _mcrA-_targeted (B) T-RFLP analysis of a soil archaeal community. Amplicons were generated from identical soil DNA templates with various PCR annealing temperatures and analyzed by T-RFLP fingerprinting. RAF, relative amplicon frequency.
FIG. 2.
Electropherograms of T-RFLP analysis of defined template mixtures of M. bryantii (MB), M. concilii (MX), M. hungatei (MP), and M. jannaschii (MC) pure-culture DNA. Shown is a series with increasing M. bryantii template ratios from 25 to 78.6% of SSU rRNA genes (A to D). T-RF lengths of characteristic peaks are indicated in parentheses (in base pairs). RFU, relative fluorescence units.
FIG. 3.
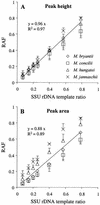
Relationship between SSU rDNA template ratios and relative amplicon frequencies as determined by T-RFLP analysis of defined template mixtures. Relative amplicon frequencies (RAF) were determined by fluorescence peak height analysis (A) or peak area integration (B). Lines indicate best linear fit for combined data sets of each graph, and slopes are given. Average values of separately prepared defined template mixtures are plotted (± standard deviation, n = 3).
FIG. 4.
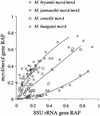
Relationship between SSU rDNA and mcrA/mrtA gene relative amplicon frequencies (RAF) as determined by T-RFLP analysis of defined template mixtures. Lines indicate estimated fits.
Similar articles
- Molecular analyses of methyl-coenzyme M reductase alpha-subunit (mcrA) genes in rice field soil and enrichment cultures reveal the methanogenic phenotype of a novel archaeal lineage.
Lueders T, Chin KJ, Conrad R, Friedrich M. Lueders T, et al. Environ Microbiol. 2001 Mar;3(3):194-204. doi: 10.1046/j.1462-2920.2001.00179.x. Environ Microbiol. 2001. PMID: 11321536 - Investigation of the methanogen population structure and activity in a brackish lake sediment.
Banning N, Brock F, Fry JC, Parkes RJ, Hornibrook ER, Weightman AJ. Banning N, et al. Environ Microbiol. 2005 Jul;7(7):947-60. doi: 10.1111/j.1462-2920.2004.00766.x. Environ Microbiol. 2005. PMID: 15946291 - An evaluation of terminal-restriction fragment length polymorphism (T-RFLP) analysis for the study of microbial community structure and dynamics.
Osborn AM, Moore ER, Timmis KN. Osborn AM, et al. Environ Microbiol. 2000 Feb;2(1):39-50. doi: 10.1046/j.1462-2920.2000.00081.x. Environ Microbiol. 2000. PMID: 11243261 - Technicalities and Glitches of Terminal Restriction Fragment Length Polymorphism (T-RFLP).
Prakash O, Pandey PK, Kulkarni GJ, Mahale KN, Shouche YS. Prakash O, et al. Indian J Microbiol. 2014 Sep;54(3):255-61. doi: 10.1007/s12088-014-0461-0. Epub 2014 Mar 9. Indian J Microbiol. 2014. PMID: 24891731 Free PMC article. Review. - Methanogens: reevaluation of a unique biological group.
Balch WE, Fox GE, Magrum LJ, Woese CR, Wolfe RS. Balch WE, et al. Microbiol Rev. 1979 Jun;43(2):260-96. doi: 10.1128/mr.43.2.260-296.1979. Microbiol Rev. 1979. PMID: 390357 Free PMC article. Review. No abstract available.
Cited by
- Current Methods, Common Practices, and Perspectives in Tracking and Monitoring Bioinoculants in Soil.
Manfredini A, Malusà E, Costa C, Pallottino F, Mocali S, Pinzari F, Canfora L. Manfredini A, et al. Front Microbiol. 2021 Aug 31;12:698491. doi: 10.3389/fmicb.2021.698491. eCollection 2021. Front Microbiol. 2021. PMID: 34531836 Free PMC article. Review. - Methods for exploring the faecal microbiome of premature infants: a review.
Westaway JAF, Huerlimann R, Miller CM, Kandasamy Y, Norton R, Rudd D. Westaway JAF, et al. Matern Health Neonatol Perinatol. 2021 Mar 8;7(1):11. doi: 10.1186/s40748-021-00131-9. Matern Health Neonatol Perinatol. 2021. PMID: 33685524 Free PMC article. Review. - Seasonal Dynamics of Abundance, Structure, and Diversity of Methanogens and Methanotrophs in Lake Sediments.
Lyautey E, Billard E, Tissot N, Jacquet S, Domaizon I. Lyautey E, et al. Microb Ecol. 2021 Oct;82(3):559-571. doi: 10.1007/s00248-021-01689-9. Epub 2021 Feb 4. Microb Ecol. 2021. PMID: 33538855 - Scorpion primer PCR analysis for genotyping of allele variants of thiopurine s‑methyltransferase*3.
Yao P, Qu XM, Ren S, Ren XD, Su N, Zhao N, Wang L, Cheng L, Weng BB, Sun FJ, Huang Q. Yao P, et al. Mol Med Rep. 2020 Sep;22(3):1994-2002. doi: 10.3892/mmr.2020.11283. Epub 2020 Jun 26. Mol Med Rep. 2020. PMID: 32705177 Free PMC article.
References
- Avaniss-Aghajani, E., K. Jones, D. Chapman, and C. Brunk. 1994. A molecular technique for identification of bacteria with small subunit ribosomal RNA sequences. BioTechniques 17:144-149. - PubMed
- Berthelet, M., L. G. Whyte, and C. W. Greer. 1996. Rapid, direct extraction of DNA from soils for PCR analysis with polyvinylpolypyrrolidone spin columns. FEMS Microbiol. Lett. 138:17-22. - PubMed
- Braker, G., H. L. Ayala-del-Rio, A. H. Devol, A. Fesefeldt, and J. M. Tiedje. 2001. Community structure of denitrifiers, Bacteria, and Archaea along redox gradients in pacific northwest marine sediments by terminal restriction fragment length polymorphism analysis of amplified nitrite reductase (nirS) and 16S rRNA genes. Appl. Environ. Microbiol. 67:1893-1901. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources