Mechanism of histone lysine methyl transfer revealed by the structure of SET7/9-AdoMet - PubMed (original) (raw)
Mechanism of histone lysine methyl transfer revealed by the structure of SET7/9-AdoMet
Taewoo Kwon et al. EMBO J. 2003.
Abstract
The methylation of lysine residues of histones plays a pivotal role in the regulation of chromatin structure and gene expression. Here, we report two crystal structures of SET7/9, a histone methyltransferase (HMTase) that transfers methyl groups to Lys4 of histone H3, in complex with S-adenosyl-L-methionine (AdoMet) determined at 1.7 and 2.3 A resolution. The structures reveal an active site consisting of: (i) a binding pocket between the SET domain and a c-SET helix where an AdoMet molecule in an unusual conformation binds; (ii) a narrow substrate-specific channel that only unmethylated lysine residues can access; and (iii) a catalytic tyrosine residue. The methyl group of AdoMet is directed to the narrow channel where a substrate lysine enters from the opposite side. We demonstrate that SET7/9 can transfer two but not three methyl groups to unmodified Lys4 of H3 without substrate dissociation. The unusual features of the SET domain-containing HMTase discriminate between the un- and methylated lysine substrate, and the methylation sites for the histone H3 tail.
Figures
Fig. 1. (A) Stereo diagram of the 1.7 Å resolution electron density map around the methyl group of AdoMet. The |_F_o – _F_c| simulated annealing omit map contoured at the 3σ level, with AdoMet and neighbouring residues omitted for map calculation. The refined 1.7 Å structure is superimposed on the map. (B) Sequence alignment of SET7/9, human SUV39H1 and the yeast homologue Clr4. Residues identical between SET7/9, SUV39H1 and Clr4 are highlighted in yellow. The bar graph below the sequence alignment indicates the degree of identity among 11 SET domain-containing HMTases: Hs SET9, Mm Suv39h1, Hs Suv39H1, Dm Su(var)3–9, Hs G9a, Sc SET1, Sp Clr4, Nc dim-5, Hs PR-SET7, Sc SET2, Mm ESET and Hs ESET. Insertions in SUV39H1 and Clr4 sequences are dropped below the alignment with a vertical line for clarity. Loops that form the active site pocket are highlighted in orange, and residues that make up the pocket walls and active site, and those that contact AdoMet are indicated.
Fig. 1. (A) Stereo diagram of the 1.7 Å resolution electron density map around the methyl group of AdoMet. The |_F_o – _F_c| simulated annealing omit map contoured at the 3σ level, with AdoMet and neighbouring residues omitted for map calculation. The refined 1.7 Å structure is superimposed on the map. (B) Sequence alignment of SET7/9, human SUV39H1 and the yeast homologue Clr4. Residues identical between SET7/9, SUV39H1 and Clr4 are highlighted in yellow. The bar graph below the sequence alignment indicates the degree of identity among 11 SET domain-containing HMTases: Hs SET9, Mm Suv39h1, Hs Suv39H1, Dm Su(var)3–9, Hs G9a, Sc SET1, Sp Clr4, Nc dim-5, Hs PR-SET7, Sc SET2, Mm ESET and Hs ESET. Insertions in SUV39H1 and Clr4 sequences are dropped below the alignment with a vertical line for clarity. Loops that form the active site pocket are highlighted in orange, and residues that make up the pocket walls and active site, and those that contact AdoMet are indicated.
Fig. 2. Overall structure of SET7/9–AdoMet. (A) The 2.3 Å SET7/9L structure is shown in a ribbon representation. A bound AdoMet molecule is shown in a ball-and-stick model (oxygen, red; carbon, cyan; nitrogen, blue; and sulfur, green). The n-, SET and c-SET regions in the C-terminal domain are coloured in yellow, red and green, respectively. (B) Topological diagrams of secondary structure elements. Loops forming the active site pocket are in red. The AdoMet-binding site is indicated. The colour scheme is identical to that in (A). (C) Stereo diagram showing an extensive van der Waals contacts network between the SET and c-SET region. The AdoMet is coloured in cyan, and the residues from SET and c-SET domains are in yellow. Other atoms are shown in the same colour as in (A).
Fig. 2. Overall structure of SET7/9–AdoMet. (A) The 2.3 Å SET7/9L structure is shown in a ribbon representation. A bound AdoMet molecule is shown in a ball-and-stick model (oxygen, red; carbon, cyan; nitrogen, blue; and sulfur, green). The n-, SET and c-SET regions in the C-terminal domain are coloured in yellow, red and green, respectively. (B) Topological diagrams of secondary structure elements. Loops forming the active site pocket are in red. The AdoMet-binding site is indicated. The colour scheme is identical to that in (A). (C) Stereo diagram showing an extensive van der Waals contacts network between the SET and c-SET region. The AdoMet is coloured in cyan, and the residues from SET and c-SET domains are in yellow. Other atoms are shown in the same colour as in (A).
Fig. 2. Overall structure of SET7/9–AdoMet. (A) The 2.3 Å SET7/9L structure is shown in a ribbon representation. A bound AdoMet molecule is shown in a ball-and-stick model (oxygen, red; carbon, cyan; nitrogen, blue; and sulfur, green). The n-, SET and c-SET regions in the C-terminal domain are coloured in yellow, red and green, respectively. (B) Topological diagrams of secondary structure elements. Loops forming the active site pocket are in red. The AdoMet-binding site is indicated. The colour scheme is identical to that in (A). (C) Stereo diagram showing an extensive van der Waals contacts network between the SET and c-SET region. The AdoMet is coloured in cyan, and the residues from SET and c-SET domains are in yellow. Other atoms are shown in the same colour as in (A).
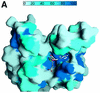
Fig. 3. The AdoMet-binding pocket. (A) Surface representation of the AdoMet-binding pocket. The surface is coloured according to the residue identity as in Figure 1B, highlighting the conservation of residues that make up the AdoMet-binding site. The AdoMet molecule is shown in a bond representation. The orientation of the figure is similar to that in (B). (B) The AdoMet-binding pocket in SET7/9S. Key residues discussed in the text are shown in a ball-and-stick model. The AdoMet molecule is shown with same colour scheme as in (A), except that carbon atoms are shown in cyan. (C) Stereo diagram showing the conformations of AdoMet molecules. Three AdoMet molecules, extended (red), intermediate (blue) and compact (yellow) forms, are superimposed. The extended AdoMet molecule was drawn using AdoMet bound to isoflavone-_O_-methyltransferase [Protein data bank (PDB) identifiaction code: 1FPX], and the intermediate form was from AdoMet bound to methionine repressor protein (Metj; PDB identification code: 1CMA). (D) A schematic drawing showing direct interactions between SET7/9 and AdoMet. Hydrogen bonds are shown by dashed lines. An arc next to the residue name indicates that the amino acid is involved in a van der Waals interaction with AdoMet. The residues coloured in blue indicate side chain interactions, while those in red are involved in backbone interactions.
Fig. 3. The AdoMet-binding pocket. (A) Surface representation of the AdoMet-binding pocket. The surface is coloured according to the residue identity as in Figure 1B, highlighting the conservation of residues that make up the AdoMet-binding site. The AdoMet molecule is shown in a bond representation. The orientation of the figure is similar to that in (B). (B) The AdoMet-binding pocket in SET7/9S. Key residues discussed in the text are shown in a ball-and-stick model. The AdoMet molecule is shown with same colour scheme as in (A), except that carbon atoms are shown in cyan. (C) Stereo diagram showing the conformations of AdoMet molecules. Three AdoMet molecules, extended (red), intermediate (blue) and compact (yellow) forms, are superimposed. The extended AdoMet molecule was drawn using AdoMet bound to isoflavone-_O_-methyltransferase [Protein data bank (PDB) identifiaction code: 1FPX], and the intermediate form was from AdoMet bound to methionine repressor protein (Metj; PDB identification code: 1CMA). (D) A schematic drawing showing direct interactions between SET7/9 and AdoMet. Hydrogen bonds are shown by dashed lines. An arc next to the residue name indicates that the amino acid is involved in a van der Waals interaction with AdoMet. The residues coloured in blue indicate side chain interactions, while those in red are involved in backbone interactions.
Fig. 3. The AdoMet-binding pocket. (A) Surface representation of the AdoMet-binding pocket. The surface is coloured according to the residue identity as in Figure 1B, highlighting the conservation of residues that make up the AdoMet-binding site. The AdoMet molecule is shown in a bond representation. The orientation of the figure is similar to that in (B). (B) The AdoMet-binding pocket in SET7/9S. Key residues discussed in the text are shown in a ball-and-stick model. The AdoMet molecule is shown with same colour scheme as in (A), except that carbon atoms are shown in cyan. (C) Stereo diagram showing the conformations of AdoMet molecules. Three AdoMet molecules, extended (red), intermediate (blue) and compact (yellow) forms, are superimposed. The extended AdoMet molecule was drawn using AdoMet bound to isoflavone-_O_-methyltransferase [Protein data bank (PDB) identifiaction code: 1FPX], and the intermediate form was from AdoMet bound to methionine repressor protein (Metj; PDB identification code: 1CMA). (D) A schematic drawing showing direct interactions between SET7/9 and AdoMet. Hydrogen bonds are shown by dashed lines. An arc next to the residue name indicates that the amino acid is involved in a van der Waals interaction with AdoMet. The residues coloured in blue indicate side chain interactions, while those in red are involved in backbone interactions.
Fig. 3. The AdoMet-binding pocket. (A) Surface representation of the AdoMet-binding pocket. The surface is coloured according to the residue identity as in Figure 1B, highlighting the conservation of residues that make up the AdoMet-binding site. The AdoMet molecule is shown in a bond representation. The orientation of the figure is similar to that in (B). (B) The AdoMet-binding pocket in SET7/9S. Key residues discussed in the text are shown in a ball-and-stick model. The AdoMet molecule is shown with same colour scheme as in (A), except that carbon atoms are shown in cyan. (C) Stereo diagram showing the conformations of AdoMet molecules. Three AdoMet molecules, extended (red), intermediate (blue) and compact (yellow) forms, are superimposed. The extended AdoMet molecule was drawn using AdoMet bound to isoflavone-_O_-methyltransferase [Protein data bank (PDB) identifiaction code: 1FPX], and the intermediate form was from AdoMet bound to methionine repressor protein (Metj; PDB identification code: 1CMA). (D) A schematic drawing showing direct interactions between SET7/9 and AdoMet. Hydrogen bonds are shown by dashed lines. An arc next to the residue name indicates that the amino acid is involved in a van der Waals interaction with AdoMet. The residues coloured in blue indicate side chain interactions, while those in red are involved in backbone interactions.
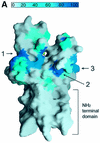
Fig. 4. A channel at the active site pocket. (A) A surface representation showing the location of the putative lysine-binding channel and a conserved shallow groove for the substrate-binding site (indicated by an arrow). See the text for sites 1, 2 and 3. The colouring scheme is identical to that in Figure 3A. AdoMet is shown in a bond model. The N-terminal domain is not coloured since it is not homologous to those of other SET-containing HMTases (Figure 1B). (B) A diagram showing the substrate-specific channel in SET7/9S. Key residues discussed in the text are shown in a ball-and-stick model. The AdoMet molecule is shown with the same colour scheme as in Figure 3B. (C) SET7/9 methylates unmethylated H3 peptide but cannot add methyl group(s) to already methylated peptide. An N-terminal peptide with amino acids 1–8 of unmodified, mono- or dimethylated H3-K4 was used for the assay of SET7/9 HMTase activity. The full-length SET7/9 was used for the assay. (D) Methylation specificity of SET7/9. Histone H3 (Roche) was methylated by SET7/9. The reaction products were resolved by SDS–PAGE, blotted to nitrocellulose and probed with either a H3-K4 mono- or dimethyl antibody, or a H3-K4 trimethyl antibody as indicated. The H3-K4, K9 trimethyl antibody also gave the same result as that of the H3-K4 trimethyl antibody (data not shown).
Fig. 4. A channel at the active site pocket. (A) A surface representation showing the location of the putative lysine-binding channel and a conserved shallow groove for the substrate-binding site (indicated by an arrow). See the text for sites 1, 2 and 3. The colouring scheme is identical to that in Figure 3A. AdoMet is shown in a bond model. The N-terminal domain is not coloured since it is not homologous to those of other SET-containing HMTases (Figure 1B). (B) A diagram showing the substrate-specific channel in SET7/9S. Key residues discussed in the text are shown in a ball-and-stick model. The AdoMet molecule is shown with the same colour scheme as in Figure 3B. (C) SET7/9 methylates unmethylated H3 peptide but cannot add methyl group(s) to already methylated peptide. An N-terminal peptide with amino acids 1–8 of unmodified, mono- or dimethylated H3-K4 was used for the assay of SET7/9 HMTase activity. The full-length SET7/9 was used for the assay. (D) Methylation specificity of SET7/9. Histone H3 (Roche) was methylated by SET7/9. The reaction products were resolved by SDS–PAGE, blotted to nitrocellulose and probed with either a H3-K4 mono- or dimethyl antibody, or a H3-K4 trimethyl antibody as indicated. The H3-K4, K9 trimethyl antibody also gave the same result as that of the H3-K4 trimethyl antibody (data not shown).
Fig. 4. A channel at the active site pocket. (A) A surface representation showing the location of the putative lysine-binding channel and a conserved shallow groove for the substrate-binding site (indicated by an arrow). See the text for sites 1, 2 and 3. The colouring scheme is identical to that in Figure 3A. AdoMet is shown in a bond model. The N-terminal domain is not coloured since it is not homologous to those of other SET-containing HMTases (Figure 1B). (B) A diagram showing the substrate-specific channel in SET7/9S. Key residues discussed in the text are shown in a ball-and-stick model. The AdoMet molecule is shown with the same colour scheme as in Figure 3B. (C) SET7/9 methylates unmethylated H3 peptide but cannot add methyl group(s) to already methylated peptide. An N-terminal peptide with amino acids 1–8 of unmodified, mono- or dimethylated H3-K4 was used for the assay of SET7/9 HMTase activity. The full-length SET7/9 was used for the assay. (D) Methylation specificity of SET7/9. Histone H3 (Roche) was methylated by SET7/9. The reaction products were resolved by SDS–PAGE, blotted to nitrocellulose and probed with either a H3-K4 mono- or dimethyl antibody, or a H3-K4 trimethyl antibody as indicated. The H3-K4, K9 trimethyl antibody also gave the same result as that of the H3-K4 trimethyl antibody (data not shown).
Fig. 5. (A) Stereo diagram showing the active site region in SET7/9S. Eight residues discussed in the text along with interacting residues are shown in a ball-and-stick model. (B) HMTase activities of the SET7/9 point mutants. Reactions contained wild-type or mutant SET7/9, 1 µM AdoMet and 1 µg of H3 from calf thymus. Assays were performed in triplicate.
Fig. 5. (A) Stereo diagram showing the active site region in SET7/9S. Eight residues discussed in the text along with interacting residues are shown in a ball-and-stick model. (B) HMTase activities of the SET7/9 point mutants. Reactions contained wild-type or mutant SET7/9, 1 µM AdoMet and 1 µg of H3 from calf thymus. Assays were performed in triplicate.
Similar articles
- Catalytic properties and kinetic mechanism of human recombinant Lys-9 histone H3 methyltransferase SUV39H1: participation of the chromodomain in enzymatic catalysis.
Chin HG, Patnaik D, Estève PO, Jacobsen SE, Pradhan S. Chin HG, et al. Biochemistry. 2006 Mar 14;45(10):3272-84. doi: 10.1021/bi051997r. Biochemistry. 2006. PMID: 16519522 - Mechanism of histone methylation catalyzed by protein lysine methyltransferase SET7/9 and origin of product specificity.
Guo HB, Guo H. Guo HB, et al. Proc Natl Acad Sci U S A. 2007 May 22;104(21):8797-802. doi: 10.1073/pnas.0702981104. Epub 2007 May 15. Proc Natl Acad Sci U S A. 2007. PMID: 17517655 Free PMC article. - Structure and catalytic mechanism of the human histone methyltransferase SET7/9.
Xiao B, Jing C, Wilson JR, Walker PA, Vasisht N, Kelly G, Howell S, Taylor IA, Blackburn GM, Gamblin SJ. Xiao B, et al. Nature. 2003 Feb 6;421(6923):652-6. doi: 10.1038/nature01378. Epub 2003 Jan 22. Nature. 2003. PMID: 12540855 - SET domain protein lysine methyltransferases: Structure, specificity and catalysis.
Qian C, Zhou MM. Qian C, et al. Cell Mol Life Sci. 2006 Dec;63(23):2755-63. doi: 10.1007/s00018-006-6274-5. Cell Mol Life Sci. 2006. PMID: 17013555 Free PMC article. Review. - Cracking the histone code: one, two, three methyls, you're out!
Dutnall RN. Dutnall RN. Mol Cell. 2003 Jul;12(1):3-4. doi: 10.1016/s1097-2765(03)00282-x. Mol Cell. 2003. PMID: 12887887 Review.
Cited by
- Substrate docking-mediated specific and efficient lysine methylation by the SET domain-containing histone methyltransferase SETD7.
Liu H, Li Z, Yang Q, Liu W, Wan J, Li J, Zhang M. Liu H, et al. J Biol Chem. 2019 Sep 6;294(36):13355-13365. doi: 10.1074/jbc.RA119.009630. Epub 2019 Jul 19. J Biol Chem. 2019. PMID: 31324717 Free PMC article. - Enzymatic mechanism and product specificity of SET-domain protein lysine methyltransferases.
Zhang X, Bruice TC. Zhang X, et al. Proc Natl Acad Sci U S A. 2008 Apr 15;105(15):5728-32. doi: 10.1073/pnas.0801788105. Epub 2008 Apr 7. Proc Natl Acad Sci U S A. 2008. PMID: 18391193 Free PMC article. - Chemical mechanisms of histone lysine and arginine modifications.
Smith BC, Denu JM. Smith BC, et al. Biochim Biophys Acta. 2009 Jan;1789(1):45-57. doi: 10.1016/j.bbagrm.2008.06.005. Epub 2008 Jun 14. Biochim Biophys Acta. 2009. PMID: 18603028 Free PMC article. Review. - SET7/9 inhibits oncogenic activities through regulation of Gli-1 expression in breast cancer.
Song Y, Zhang J, Tian T, Fu X, Wang W, Li S, Shi T, Suo A, Ruan Z, Guo H, Yao Y. Song Y, et al. Tumour Biol. 2016 Jul;37(7):9311-22. doi: 10.1007/s13277-016-4822-7. Epub 2016 Jan 16. Tumour Biol. 2016. PMID: 26779630 - Catalytic and functional roles of conserved amino acids in the SET domain of the S. cerevisiae lysine methyltransferase Set1.
Williamson K, Schneider V, Jordan RA, Mueller JE, Henderson Pozzi M, Bryk M. Williamson K, et al. PLoS One. 2013;8(3):e57974. doi: 10.1371/journal.pone.0057974. Epub 2013 Mar 1. PLoS One. 2013. PMID: 23469257 Free PMC article.
References
- Bannister A.J., Zegerman,P., Partridge,J.F., Miska,E.A., Thomas,J.O., Allshire,R.C. and Kouzarides,T. (2001) Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature, 410, 120–124. - PubMed
- Boggs B.A., Cheung,P., Heard,E., Spector,D.L., Chinault,A.C. and Allis,C.D. (2002) Differentially methylated forms of histone H3 show unique association patterns with inactive human X chromosomes. Nat. Genet., 30, 73–76. - PubMed
- Brünger A.T. et al. (1998) Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. - PubMed
- Byvoet P., Shepherd,G.R., Hardin,J.M. and Noland,B.J. (1972) The distribution and turnover of labeled methyl groups in histone fractions of cultured mammalian cells. Arch. Biochem. Biophys., 148, 558–567. - PubMed
- CCP4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases