Excess SeqA prolongs sequestration of oriC and delays nucleoid segregation and cell division - PubMed (original) (raw)
Excess SeqA prolongs sequestration of oriC and delays nucleoid segregation and cell division
Trond Bach et al. EMBO J. 2003.
Abstract
Following initiation of chromosomal replication in Escherichia coli, newly initiated origins (oriCs) are prevented from further initiations by a mechanism termed sequestration. During the sequestration period (which lasts about one-third of a cell cycle), the origins remain hemimethylated. The SeqA protein binds hemimethylated oriC in vitro. In vivo, the absence of SeqA causes overinitiation and strongly reduces the duration of hemimethylation. The pattern of immunostained SeqA complexes in vivo suggests that SeqA has a role in organizing hemimethylated DNA at the replication forks. We have examined the effects of overexpressing SeqA under different cellular conditions. Our data demonstrate that excess SeqA significantly increases the time oriC is hemimethylated following initiation of replication. In some cells, sequestration continued for more than one generation and resulted in inhibition of primary initiation. SeqA overproduction also interfered with the segregation of sister nucleoids and caused a delay in cell division. These results suggest that SeqA's function in regulation of replication initiation is linked to chromosome segregation and possibly cell division.
Figures
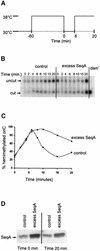
Fig. 1. Overexpression of SeqA protein extends the period of hemimethylation at oriC. (A) Schematic representation of the temperature shifts at the indicated times during the experiment. (B) Exponentially growing cultures (30°C) of MG1655 dnaC2 cells harbouring pFH2102 (control) or pMAK7 (excess SeqA) were synchronized with respect to initiation for 1 h at 38°C before shifting the temperature to 30°C. IPTG (0.5 mM) was added to the cultures three mass doublings prior to synchronized initiation. Chromosomal DNA was extracted at the indicated times after returning the cultures to 30°C. The DNA was digested with _Xho_I and _Hph_I, electrophoresed and hybridized to an oriC probe. The positions of cut and uncut fragments are indicated by arrows. The rightmost lane contains DNA isolated from a _dam_– strain which reveals the position of the cut fragment. (C) Quantification of the amount of hemimethylated oriC in (B). Squares represent _dnaC_2/pFH2102 and triangles _dnaC_2/pMAK7. The percentage of hemimethylated oriC is twice the percentage of the cut fragment in (B) since 50% of the hemimethylated fragments are digested. Time indicates minutes from shift to 30°C after 1 h synchronization at 38°C. (D) Western analyses of samples taken from the cultures at 0 and 20 min after returning the cultures to 30°C. A 1 µg aliquot of total protein of each sample was loaded.
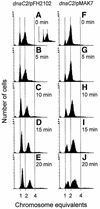
Fig. 2. DNA content histograms of dnaC2/pFH2102 (A–E), dnaC2/pMAK7 (F–J) and dnaC2/pSF19 (A, small panel). Samples for flow cytometry of dnaC2/pFH2102 and dnaC2/pMAK7 cells were collected during the temperature shift experiment in Figure 1 at the times indicated. In an identical control experiment, dnaC2/pSF19 cells were harvested for flow cytometry, at the time indicated.
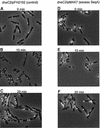
Fig. 3. Hoechst-stained CM735/pFH2102 (A) and CM735/pMAK7 cells (B–D). Cells were grown exponentially in glucose-CAA medium and then incubated with cephalexin (15 µg/ml) for two generations and with chloramphenicol (200 µg/ml) for 15 min. The CM735/pFH2102 (control) culture was incubated with 0.5 mM IPTG, and the CM735/pMAK7 cultures with 0, 0.05 or 0.5 mM IPTG, for three generations prior to sample harvesting.
Fig. 4. Immunofluorescent SeqA foci in _dnaC_2/pFH2102 (A–C) and _dnaC_2/pMAK7 (D–F) cells. Cells were from the same samples as those analysed by flow cytometry in Figure 2 (0, 10 and 20 min).
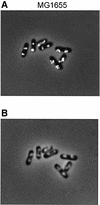
Fig. 5. Immunofluorescent SeqA foci (A) and Hoechst-stained nucleoids (B) in the same cells. Wild-type (MG1655) cells were grown exponentially in glucose-CAA medium to OD450 = 0.15. Samples were fixed in ethanol and subjected to immunofluorescence microscopy.
Similar articles
- Competition between the replication initiator DnaA and the sequestration factor SeqA for binding to the hemimethylated chromosomal origin of E. coli in vitro.
Taghbalout A, Landoulsi A, Kern R, Yamazoe M, Hiraga S, Holland B, Kohiyama M, Malki A. Taghbalout A, et al. Genes Cells. 2000 Nov;5(11):873-884. doi: 10.1046/j.1365-2443.2000.00380.x. Genes Cells. 2000. PMID: 11122375 - SeqA, the Escherichia coli origin sequestration protein, can regulate the replication of the pBR322 plasmid.
Douraid D, Ahmed L. Douraid D, et al. Plasmid. 2011 Jan;65(1):15-9. doi: 10.1016/j.plasmid.2010.09.003. Epub 2010 Sep 25. Plasmid. 2011. PMID: 20875449 - Escherichia coli SeqA protein affects DNA topology and inhibits open complex formation at oriC.
Torheim NK, Skarstad K. Torheim NK, et al. EMBO J. 1999 Sep 1;18(17):4882-8. doi: 10.1093/emboj/18.17.4882. EMBO J. 1999. PMID: 10469666 Free PMC article. - The Escherichia coli SeqA protein.
Waldminghaus T, Skarstad K. Waldminghaus T, et al. Plasmid. 2009 May;61(3):141-50. doi: 10.1016/j.plasmid.2009.02.004. Epub 2009 Feb 28. Plasmid. 2009. PMID: 19254745 Review. - Partitioning of the Escherichia coli chromosome: superhelicity and condensation.
Nordström K, Dasgupta S. Nordström K, et al. Biochimie. 2001 Jan;83(1):41-8. doi: 10.1016/s0300-9084(00)01204-9. Biochimie. 2001. PMID: 11254973 Review.
Cited by
- Multiple DNA Binding Proteins Contribute to Timing of Chromosome Replication in E. coli.
Riber L, Frimodt-Møller J, Charbon G, Løbner-Olesen A. Riber L, et al. Front Mol Biosci. 2016 Jun 28;3:29. doi: 10.3389/fmolb.2016.00029. eCollection 2016. Front Mol Biosci. 2016. PMID: 27446932 Free PMC article. Review. - Regulation of the replication cycle: conserved and diverse regulatory systems for DnaA and oriC.
Katayama T, Ozaki S, Keyamura K, Fujimitsu K. Katayama T, et al. Nat Rev Microbiol. 2010 Mar;8(3):163-70. doi: 10.1038/nrmicro2314. Nat Rev Microbiol. 2010. PMID: 20157337 - The GATC-binding protein SeqA is required for bile resistance and virulence in Salmonella enterica serovar typhimurium.
Prieto AI, Jakomin M, Segura I, Pucciarelli MG, Ramos-Morales F, García-Del Portillo F, Casadesús J. Prieto AI, et al. J Bacteriol. 2007 Dec;189(23):8496-502. doi: 10.1128/JB.01156-07. Epub 2007 Sep 28. J Bacteriol. 2007. PMID: 17905993 Free PMC article. - Mutant DnaAs of Escherichia coli that are refractory to negative control.
Chodavarapu S, Felczak MM, Simmons LA, Murillo A, Kaguni JM. Chodavarapu S, et al. Nucleic Acids Res. 2013 Dec;41(22):10254-67. doi: 10.1093/nar/gkt774. Epub 2013 Aug 29. Nucleic Acids Res. 2013. PMID: 23990329 Free PMC article. - 60 Years of Studies into the Initiation of Chromosome Replication in Bacteria.
Herrick J, Norris V, Kohiyama M. Herrick J, et al. Biomolecules. 2025 Feb 1;15(2):203. doi: 10.3390/biom15020203. Biomolecules. 2025. PMID: 40001506 Free PMC article. Review.
References
- Allen G.-C. Jr and Kornberg,A. (1991) Fine balance in the regulation of DnaB helicase by DnaC protein in replication in Escherichia coli. J. Biol. Chem., 266, 22096–22101. - PubMed
- Boye E. and Lobner-Olesen,A. (1990) The role of dam methyltransferase in the control of DNA replication in E.coli. Cell, 62, 981–989. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases