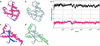Insufficiently dehydrated hydrogen bonds as determinants of protein interactions - PubMed (original) (raw)
Insufficiently dehydrated hydrogen bonds as determinants of protein interactions
Ariel Fernández et al. Proc Natl Acad Sci U S A. 2003.
Abstract
The prediction of binding sites and the understanding of interfaces associated with protein complexation remains an open problem in molecular biophysics. This work shows that a crucial factor in predicting and rationalizing protein-protein interfaces can be inferred by assessing the extent of intramolecular desolvation of backbone hydrogen bonds in monomeric structures. Our statistical analysis of native structures shows that, in the majority of soluble proteins, most backbone hydrogen bonds are thoroughly wrapped intramolecularly by nonpolar groups except for a few ones. These latter underwrapped hydrogen bonds may be dramatically stabilized by removal of water. This fact implies that packing defects are "sticky" in a way that decisively contributes to determining the binding sites for proteins, as an examination of numerous complexes demonstrates.
Figures
Figure 1
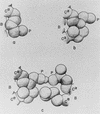
Schematic representation (5) of various hydrophobic interactions of a polar side chain with its surroundings. B, backbone; P, polar head; Cα, α-carbon. (a) Interaction of a lysine side chain with the backbone. (b) Interaction of a lysine side chain with a nearby isoleucine side chain. (c) Interaction of two polar side chains (lysine and glutamic acid), engaged in hydrogen bonding, with two nonpolar side chains (isoleucine and leucine, respectively). A hydrophobic interaction is also formed between the two nonpolar side chains. [Reproduced with permission from ref. (Biopolymers copyright 1963, copyright owner as specified in Biopolymers).]
Figure 2
(a_–_d) Ribbon structure and backbone hydrogen-bond pattern for Hb β-subunit (PDB ID code 1bz0, a and b) and human prion protein (PDB ID code 1qm0, c and d). A dark-gray series of virtual bonds joining consecutive α-carbons represents the backbone in b and d. The sufficiently wrapped amide-carbonyl hydrogen bonds are represented by light-gray thin lines joining the α-carbons of paired residues, and the underwrapped hydrogen bonds are represented by green lines (b and d). Time-dependent average extent ρ = ρ(t) of backbone hydrogen-bond dehydration (black plot), and dispersion σ = σ(t) (red plot) over all backbone hydrogen bonds appearing along a time sequence of chain conformations extracted from the Duan–Kollman trajectory (21) are shown (e).
Figure 3
Three selected complexes and separated binding partners for the HIV-1 protease dimer (PDB ID code 1a30, a and b), colicin + ligand (PDB ID code 1emv, c and d), and CheY complex (PDB ID code 1fqw, e and f). The binding partners are represented by blue and red virtual-bond backbone chains; the hydrophobic residues containing >1 carbonaceous group are denoted as α-carbon spheres: gray if the residue is >60% buried and yellow otherwise. The backbone hydrogen bonds are indicated as lines joining α-carbons: gray if the bond is sufficiently dehydrated and green if it is a UDHB. A thin blue line joining an α-carbon with a hydrogen-bond center indicates that a residue in one molecule is engaged in one or several intermolecular three-body correlations contributing to the dehydration of an intramolecular hydrogen bond of the binding partner. (g) Sufficiently dehydrated hydrogen bonds and UDHBs for monomeric β2-microglobulin. The segment with the highest concentration of structural defects is highlighted in red and is part of the so-called βB–βC (residues 21–33) amyloidogenic fragment generated by treatment of β2-microglobulin with Acromobacter protease (23). The ribbon picture is an aid to the eye.
Similar articles
- Functionality of wrapping defects in soluble proteins: what cannot be kept dry must be conserved.
Fernández A. Fernández A. J Mol Biol. 2004 Mar 19;337(2):477-83. doi: 10.1016/j.jmb.2004.01.050. J Mol Biol. 2004. PMID: 15003461 - The subunit interfaces of weakly associated homodimeric proteins.
Dey S, Pal A, Chakrabarti P, Janin J. Dey S, et al. J Mol Biol. 2010 Apr 23;398(1):146-60. doi: 10.1016/j.jmb.2010.02.020. Epub 2010 Feb 13. J Mol Biol. 2010. PMID: 20156457 - Hydration of protein-protein interfaces.
Rodier F, Bahadur RP, Chakrabarti P, Janin J. Rodier F, et al. Proteins. 2005 Jul 1;60(1):36-45. doi: 10.1002/prot.20478. Proteins. 2005. PMID: 15856483 - Protein wrapping: a molecular marker for association, aggregation and drug design.
Fernández A, Crespo A. Fernández A, et al. Chem Soc Rev. 2008 Nov;37(11):2373-82. doi: 10.1039/b804150b. Epub 2008 Sep 15. Chem Soc Rev. 2008. PMID: 18949110 Review. - Prediction of protein-protein interactions: unifying evolution and structure at protein interfaces.
Tuncbag N, Gursoy A, Keskin O. Tuncbag N, et al. Phys Biol. 2011 Jun;8(3):035006. doi: 10.1088/1478-3975/8/3/035006. Epub 2011 May 13. Phys Biol. 2011. PMID: 21572173 Review.
Cited by
- Structural defects and the diagnosis of amyloidogenic propensity.
Fernández A, Kardos J, Scott LR, Goto Y, Berry RS. Fernández A, et al. Proc Natl Acad Sci U S A. 2003 May 27;100(11):6446-51. doi: 10.1073/pnas.0731893100. Epub 2003 May 12. Proc Natl Acad Sci U S A. 2003. PMID: 12743379 Free PMC article. - Residue-specific side-chain packing determines the backbone dynamics of transmembrane model helices.
Quint S, Widmaier S, Minde D, Hornburg D, Langosch D, Scharnagl C. Quint S, et al. Biophys J. 2010 Oct 20;99(8):2541-9. doi: 10.1016/j.bpj.2010.08.031. Biophys J. 2010. PMID: 20959095 Free PMC article. - Simulations of mutant p53 DNA binding domains reveal a novel druggable pocket.
Pradhan MR, Siau JW, Kannan S, Nguyen MN, Ouaray Z, Kwoh CK, Lane DP, Ghadessy F, Verma CS. Pradhan MR, et al. Nucleic Acids Res. 2019 Feb 28;47(4):1637-1652. doi: 10.1093/nar/gky1314. Nucleic Acids Res. 2019. PMID: 30649466 Free PMC article. - Effect of dehydration on the aggregation kinetics of two amyloid peptides.
Mukherjee S, Chowdhury P, Gai F. Mukherjee S, et al. J Phys Chem B. 2009 Jan 15;113(2):531-5. doi: 10.1021/jp809817s. J Phys Chem B. 2009. PMID: 19132862 Free PMC article. - An in silico study of the effect of SOD1 electrostatic loop dynamics on amyloid‑like filament formation.
Healy EF, Cervantes L. Healy EF, et al. Eur Biophys J. 2016 Dec;45(8):853-859. doi: 10.1007/s00249-016-1163-9. Epub 2016 Aug 5. Eur Biophys J. 2016. PMID: 27496206 Free PMC article.
References
- Némethy G, Scheraga H A. J Phys Chem. 1962;66:1773–1789. , and erratum (1963) 67, 2888.
- Némethy G, Scheraga H A. J Chem Phys. 1962;36:3382–3400.
- Némethy G, Scheraga H A. J Chem Phys. 1962;36:3401–3417.
- Scheraga H A. J Biomol Struct Dyn. 1998;16:447–460. - PubMed
- Némethy G, Steinberg I Z, Scheraga H A. Biopolymers. 1963;1:43–69.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources