ArrayExpress--a public repository for microarray gene expression data at the EBI - PubMed (original) (raw)
. 2003 Jan 1;31(1):68-71.
doi: 10.1093/nar/gkg091.
Helen Parkinson, Ugis Sarkans, Mohammadreza Shojatalab, Jaak Vilo, Niran Abeygunawardena, Ele Holloway, Misha Kapushesky, Patrick Kemmeren, Gonzalo Garcia Lara, Ahmet Oezcimen, Philippe Rocca-Serra, Susanna-Assunta Sansone
Affiliations
- PMID: 12519949
- PMCID: PMC165538
- DOI: 10.1093/nar/gkg091
ArrayExpress--a public repository for microarray gene expression data at the EBI
Alvis Brazma et al. Nucleic Acids Res. 2003.
Abstract
ArrayExpress is a new public database of microarray gene expression data at the EBI, which is a generic gene expression database designed to hold data from all microarray platforms. ArrayExpress uses the annotation standard Minimum Information About a Microarray Experiment (MIAME) and the associated XML data exchange format Microarray Gene Expression Markup Language (MAGE-ML) and it is designed to store well annotated data in a structured way. The ArrayExpress infrastructure consists of the database itself, data submissions in MAGE-ML format or via an online submission tool MIAMExpress, online database query interface, and the Expression Profiler online analysis tool. ArrayExpress accepts three types of submission, arrays, experiments and protocols, each of these is assigned an accession number. Help on data submission and annotation is provided by the curation team. The database can be queried on parameters such as author, laboratory, organism, experiment or array types. With an increasing number of organisations adopting MAGE-ML standard, the volume of submissions to ArrayExpress is increasing rapidly. The database can be accessed at http://www.ebi.ac.uk/arrayexpress.
Figures
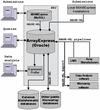
Figure 1
A representation of the dataflow, database structure and MAGE-ML based infrastructure for data sharing.
Similar articles
- ArrayExpress--a public repository for microarray gene expression data at the EBI.
Parkinson H, Sarkans U, Shojatalab M, Abeygunawardena N, Contrino S, Coulson R, Farne A, Lara GG, Holloway E, Kapushesky M, Lilja P, Mukherjee G, Oezcimen A, Rayner T, Rocca-Serra P, Sharma A, Sansone S, Brazma A. Parkinson H, et al. Nucleic Acids Res. 2005 Jan 1;33(Database issue):D553-5. doi: 10.1093/nar/gki056. Nucleic Acids Res. 2005. PMID: 15608260 Free PMC article. - ArrayExpress: a public database of gene expression data at EBI.
Rocca-Serra P, Brazma A, Parkinson H, Sarkans U, Shojatalab M, Contrino S, Vilo J, Abeygunawardena N, Mukherjee G, Holloway E, Kapushesky M, Kemmeren P, Lara GG, Oezcimen A, Sansone SA. Rocca-Serra P, et al. C R Biol. 2003 Oct-Nov;326(10-11):1075-8. doi: 10.1016/j.crvi.2003.09.026. C R Biol. 2003. PMID: 14744115 - Plant-based microarray data at the European Bioinformatics Institute. Introducing AtMIAMExpress, a submission tool for Arabidopsis gene expression data to ArrayExpress.
Mukherjee G, Abeygunawardena N, Parkinson H, Contrino S, Durinck S, Farne A, Holloway E, Lilja P, Moreau Y, Oezcimen A, Rayner T, Sharma A, Brazma A, Sarkans U, Shojatalab M. Mukherjee G, et al. Plant Physiol. 2005 Oct;139(2):632-6. doi: 10.1104/pp.105.063156. Plant Physiol. 2005. PMID: 16219923 Free PMC article. - Data storage and analysis in ArrayExpress.
Brazma A, Kapushesky M, Parkinson H, Sarkans U, Shojatalab M. Brazma A, et al. Methods Enzymol. 2006;411:370-86. doi: 10.1016/S0076-6879(06)11020-4. Methods Enzymol. 2006. PMID: 16939801 Review. - MGED standards: work in progress.
Ball CA, Brazma A. Ball CA, et al. OMICS. 2006 Summer;10(2):138-44. doi: 10.1089/omi.2006.10.138. OMICS. 2006. PMID: 16901218 Review.
Cited by
- Detection of simultaneous group effects in microRNA expression and related target gene sets.
Artmann S, Jung K, Bleckmann A, Beissbarth T. Artmann S, et al. PLoS One. 2012;7(6):e38365. doi: 10.1371/journal.pone.0038365. Epub 2012 Jun 19. PLoS One. 2012. PMID: 22723856 Free PMC article. - Probe Region Expression Estimation for RNA-Seq Data for Improved Microarray Comparability.
Uziela K, Honkela A. Uziela K, et al. PLoS One. 2015 May 12;10(5):e0126545. doi: 10.1371/journal.pone.0126545. eCollection 2015. PLoS One. 2015. PMID: 25966034 Free PMC article. - PLANEX: the plant co-expression database.
Yim WC, Yu Y, Song K, Jang CS, Lee BM. Yim WC, et al. BMC Plant Biol. 2013 May 20;13:83. doi: 10.1186/1471-2229-13-83. BMC Plant Biol. 2013. PMID: 23688397 Free PMC article. - A Semi-Automated Workflow for FAIR Maturity Indicators in the Life Sciences.
Ammar A, Bonaretti S, Winckers L, Quik J, Bakker M, Maier D, Lynch I, van Rijn J, Willighagen E. Ammar A, et al. Nanomaterials (Basel). 2020 Oct 20;10(10):2068. doi: 10.3390/nano10102068. Nanomaterials (Basel). 2020. PMID: 33092028 Free PMC article. - A Modular Repository-based Infrastructure for Simulation Model Storage and Execution Support in the Context of In Silico Oncology and In Silico Medicine.
Christodoulou NA, Tousert NE, Georgiadi EC, Argyri KD, Misichroni FD, Stamatakos GS. Christodoulou NA, et al. Cancer Inform. 2016 Oct 27;15:219-235. doi: 10.4137/CIN.S40189. eCollection 2016. Cancer Inform. 2016. PMID: 27812280 Free PMC article.
References
- The Chipping Forecast (1999) Suppl. Nature Genet., 21, 1–60.
- Brazma A., Robinson,A., Cameron,G. and Ashburner,M. (2000) One-stop shop for microarray data. Nature, 403, 699–700. - PubMed
- Brazma A., Hingamp,P., Quackenbush,J., Sherlock,G., Spellman,P., Stoeckert,C., Aach,J., Ansorge,W., Ball,C.A., Causton,H.C., Gaasterland,T., Glenisson,P., Holstege,F.C.P., Kim,I.F., Markowitz,V., Matese,J.C., Parkinson,H., Robinson,A., Sarkans,U., Schulze-Kremer,S., Stewart,J., Taylor,R., Vilo,J. and Vingron,M. (2001) Minimum information about a microarray experiment (MIAME)—toward standards for microarray data. Nature Genet., 29, 365–371. - PubMed
- Spellman P.T., Miller,M., Stewart,J., Troup,C., Sarkans,U., Chervitz,S., Bernhart,D., Sherlock,G., Ball,C., Lepage,M., Swiatek,M., Marks,W.L., Goncalves,J., Markel,S., Iordan,D., Shojatalab,M., Pizarro,A., White,J., Hubley,R., Deutsch,E., Senger,M., Aronow,B.J., Robinson,A., Bassett,D., Stoeckert,C.J.,Jr. and Brazma,A. (2002) Design and implementation of microarray gene expression markup language (MAGE-ML). Genome Biol., 3, research0046.1–0046.9. - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources