Properly folded nonstructural polyprotein directs the semliki forest virus replication complex to the endosomal compartment - PubMed (original) (raw)
Properly folded nonstructural polyprotein directs the semliki forest virus replication complex to the endosomal compartment
Anne Salonen et al. J Virol. 2003 Feb.
Abstract
The late RNA synthesis in alphavirus-infected cells, generating plus-strand RNAs, takes place on cytoplasmic vacuoles (CPVs), which are modified endosomes and lysosomes. The cytosolic surface of CPVs consists of regular membrane invaginations or spherules, which are the sites of RNA synthesis (P. Kujala, A. Ikäheimonen, N. Ehsani, H. Vihinen, P. Auvinen, and L. Kääriäinen J. Virol. 75:3873-3884, 2001). To understand how CPVs arise, we have expressed the individual Semliki Forest virus (SFV) nonstructural proteins nsP1 to nsP4 in different combinations, as well as their precursor polyprotein P1234 and its cleavage intermediates. A complex of nsPs was obtained from P123 or P1234, indicating that the precursor stage is essential for the assembly of the polymerase complex. To prevent the processing of the polyprotein and its cleavage intermediates, constructs with the mutation C478A (designated with a superscript CA) in the active site of the protease domain of nsP2 were used. Uncleaved polyproteins containing nsP1 were membrane bound and palmitoylated, and those containing nsP3 were phosphorylated, reflecting properties of authentic nsP1 and nsP3, respectively. Similarly, polyproteins containing nsP1 or nsP2 had enzymatic activities specific for the individual proteins, indicating that they were correctly folded in the precursor state. Uncleaved P12(CA) was localized almost exclusively to the plasma membrane and filopodia, like nsP1 alone, whereas P12(CA)3 and P12(CA)34 were found on cytoplasmic vesicles, some of which contained late endosomal markers. In immunoelectron microscopy these vesicles resembled CPVs in SFV-infected cells. Our results indicate that the nsP1 domain alone is responsible for the membrane association of the nonstructural polyprotein, whereas the nsP1 domain together with the nsP3 domain targets it to the intracellular vesicles.
Figures
FIG. 1.
Constructs expressing SFV nsPs and polyproteins. The proteins are shown schematically on the left. Different shadings represent cleavage products (nsPs) within the nonstructural coding region. Constructs containing inactivated nsP2 protease C478ACA produce nonprocessed polyproteins, whereas wild-type polyproteins are processed to individual nsPs (see also Fig. 2C). Recombinant adeno- and baculovirus systems as well as transfection of plasmids were utilized for expression of the proteins under study.
FIG. 2.
Expression of SFV nsPs and polyproteins. (A and B) Immunoprecipitation of SFV nsPs from transfected HeLa cells. Recombinant vaccinia virus (MVA)-infected cells were transfected with two plasmids encoding nsPs (combinations are indicated at the top) and labeled with [35S]methionine from 3 to 5 h posttransfection, followed by a 30-min chase. The postnuclear supernatant was subjected to immunoprecipitation with appropriate anti-nsP antibodies as indicated, and proteins were analyzed by SDS-PAGE in 10% gels. Expression of nsP4 often results in a double band, where the upper one represents full-length nsP4. In panel B, 0.1% bisacrylamide was used for the nsP1-nsP3 combination to enable their separation in electrophoresis. (C) Coexpression of nsP1, nsP2, nsP3, and nsP4 in transfected HeLa cells as in panels A and B. SDS-PAGE after two successive immunoprecipitations is shown. First precipitations were done with anti-nsP1 (lanes 1 and 2), anti-nsP2 (lanes 3 and 4), and anti-nsP3 (lanes 5 and 6), followed by second precipitations with a combination of anti-nsP1 and anti-nsP4 (lanes 1, 3, and 5) and anti-nsP2 and anti-nsP3 (lanes 2, 4, and 6). (D) Expression of SFV nonstructural polyproteins by the baculovirus system. Tn5 cells were infected with the indicated baculovirus-SFV polyprotein recombinants at 10 PFU/cell. Cells were harvested at 44 h after infection and subsequently lysed by boiling in Laemmli sample buffer. Samples were analyzed by SDS-PAGE in 7.5% gels and detected by immunoblotting with anti-nsP3 (lanes 1 to 7) or anti-nsP1 (lanes 8 and 9) antibodies by using the ECL procedure (Amersham Pharmacia Biotech). (E) Immunoprecipitation-recapture experiment with cleavable P1234 and P123 polyproteins. Tn5 cells were infected with baculoviruses encoding SFV P1234 and P123 at 5 PFU/cell. Cells were labeled with [35S]methionine at 42 to 44 h after infection, chased for 1 h, and collected. The P15 fraction was subjected to two successive immunoprecipitations. Samples were first immunoprecipitated with anti-nsP1 under nondenaturing conditions (lanes 1 and 4). The precipitates were denatured and subjected to a second immunoprecipitation with anti-nsP1 plus anti-nsP4 (lane 2), anti-nsP2 plus anti-nsP3 (lanes 3 and 6), or anti-nsP1 (lane 5). Analysis was done by SDS-PAGE in 10% gels. Molecular weight markers (Mw), in thousands, are shown on the left and the positions of nsPs or polyproteins are shown on the right in all panels.
FIG. 3.
Membrane association, palmitoylation, and phosphorylation of polyproteins. (A) Bac12CA3-infected Tn5 cells were labeled as for Fig. 2E. Postnuclear supernatant was subjected to flotation in a discontinuous sucrose (weight/weight) gradient consisting of 0.675 ml of 10%, 3.5 ml of 50%, and 0.625 ml of 60% sucrose on top of a 0.5-ml 67% sucrose cushion. Fractions of 0.5 ml were collected from the top and analyzed by immunoblotting with antiserum against nsP1. The sample was initially included in the 60% sucrose layer (fractions 8 and 9), and upon centrifugation, membrane-bound proteins floated to the 10 to 50% sucrose interface (fractions 2 and 3). (B) Fractions 2 and 3 were pooled, and aliquots were subjected to treatments with 50 mM Tris-HCl (pH 7.5)-100 mM NaCl (TN buffer), 1 M NaCl in TN buffer, and 50 mM Na2CO3 at pH 12. After 30 min of incubation on ice, the samples were centrifuged at 70,000 × g for 30 min. The pellet (P) and supernatant (S) fractions were immunoprecipitated with anti-nsP3 antiserum, followed by SDS-PAGE in a 7.5% gel and autoradiography. (C) Palmitoylation of SFV P12CA3. Tn5 cells were infected with Bac12CA3 and labeled at 44 h p.i. with 300 to 400 μCi of [9,10(n)-3H]palmitic acid for 8 h. Bac123- and Bac1-infected and mock-infected cells served as controls. Cells were collected, the P15 fraction was prepared, and protein expression was verified by immunoblotting (WB) (lanes 1 and 2) with anti-nsP1 antibody. Part of the P15 fraction was immunoprecipitated by anti-nsP1 antiserum. Samples were analyzed with SDS-PAGE in a 7.5% gel, and incorporated [3H]palmitate was visualized with a PhosphorImager. (D) In vivo phosphorylation of SFV P12CA34, P12CA3, and P2CA3 proteins. Tn5 cells were infected with Bac12CA34, Bac12CA3, or Bac2CA3, labeled with [32P]orthophosphate at 41 to 44 h p.i., harvested, and lysed by boiling in 1% SDS. Proteins were immunoprecipitated with anti-nsP3 antibody and analyzed with SDS-PAGE and a PhosphorImager. In all panels, positions of proteins are indicated on the right and those of molecular weight (Mw) markers, in thousands, are indicated on the left.
FIG.4
.(A) Guanylyltransferase activity of SFV polyproteins. P15 fractions from cells infected with Bac12CA34, Bac1234, Bac12CA3, Bac123, or Bac12CA were subjected to guanylyltransferase assay in the presence (+) or absence (−) of 100 μM AdoMet. Reactions were stopped by boiling with 1% SDS and followed by immunoprecipitation with anti-nsP1 antibody. Samples were analyzed by SDS-PAGE in a 7.5% gel, followed by autoradiography. Molecular weight (Mw) standards in thousands are shown on the left. (B) NTPase activity of SFV nsP2 and nonstructural polyproteins expressed by baculovirus vector, as in panel A. nsP2 expressed and purified from Escherichia coli was used as a positive control. Proteins were immunoprecipitated (IP) from the postnuclear supernatant with the antibody indicated at the bottom and incubated with [α-32P]GTP. The NTPase reaction products were analyzed with thin-layer chromatography. The positions of the reaction products are indicated with arrows. Baculo wt, baculovirus vector.
FIG. 5.
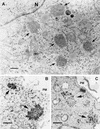
Immunolocalization analysis of SFV nonstructural polyproteins. Transfected (A, B, C, and E) or recombinant adenovirus-infected (D, F, G, and I) HeLa cells were fixed with 4% paraformaldehyde 28 h after transfection or infection. SFV-infected cells (H) (50 PFU/cell) were fixed at 5 h p.i. Cells were permeabilized and double labeled with rabbit and guinea pig antibodies against nsP1 and nsP2 (A and B), nsP1 and nsP3 (C and F), nsP2 and nsP3 (D), nsP3 and nsP4 (E), or nsP3 and lysosomal marker lamp2 (G, H, and I). Rhodamine Red X-conjugated anti-rabbit (red) and fluorescein-conjugated anti-guinea pig (green) antisera were used as secondary antibodies. Merged confocal microscopy images are shown.
FIG. 6.
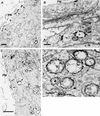
Immunogold localization of P12CA3 expressed by adenovirus vector (A and B) and nsP3 or nsP1 in SFV infection (C and D). HeLa cells were infected with adenovirus 12CA3 or SFV and fixed with PLP fixative for 2 h at 28 or 5 h p.i., respectively. Proteins were visualized by treating saponin-permeabilized cells with anti-nsP3 (A, B, and C) or anti-nsP1 (D) rabbit antiserum and nanogold-conjugated Fab fragments. P12CA3 was seen in patches at the plasma membrane (A) and on the outer surface of electron-lucent cytoplasmic vesicles (marked with an asterisk in panel B). In SFV-infected cells nsP1 and nsP3 were on the cytoplasmic side of the limiting membrane of CPVs (C and D) and in patches at the plasma membrane (C). Note that some spherules remained intact even after saponin treatment (arrows in D). Arrows indicate areas positive for immunostaining. Bars, 500 nm (A and D), 200 nm (B), and 2 μm (C). PM, plasma membrane; N, nucleus.
FIG. 7.
Aggregate-like structures of nsP3. HeLa cells were infected with adenovirus AdP3 (A and B) or with SFV (C) and fixed at 28 or 5 h p.i., respectively, using 2.5% glutaraldehyde (A) or treatment supporting the localization by the nanogold technique (B and C) as for Fig. 6. In SFV-infected cells nsP3 was also found abundantly on the cytoplasmic side of the limiting membrane of CPVs and to a lesser extent on the plasma membrane (see Fig. 6C). Putative aggresomes of nsP3 are indicated by arrows. Bars, 500 nm. PM, plasma membrane; N, nucleus.
Similar articles
- Regulation of Semliki Forest virus RNA replication: a model for the control of alphavirus pathogenesis in invertebrate hosts.
Kim KH, Rümenapf T, Strauss EG, Strauss JH. Kim KH, et al. Virology. 2004 May 20;323(1):153-63. doi: 10.1016/j.virol.2004.03.009. Virology. 2004. PMID: 15165827 - Polyprotein Processing as a Determinant for in Vitro Activity of Semliki Forest Virus Replicase.
Pietilä MK, Albulescu IC, Hemert MJV, Ahola T. Pietilä MK, et al. Viruses. 2017 Oct 7;9(10):292. doi: 10.3390/v9100292. Viruses. 2017. PMID: 28991178 Free PMC article. - Regulation of the sequential processing of Semliki Forest virus replicase polyprotein.
Vasiljeva L, Merits A, Golubtsov A, Sizemskaja V, Kääriäinen L, Ahola T. Vasiljeva L, et al. J Biol Chem. 2003 Oct 24;278(43):41636-45. doi: 10.1074/jbc.M307481200. Epub 2003 Aug 12. J Biol Chem. 2003. PMID: 12917405 - Viral RNA replication in association with cellular membranes.
Salonen A, Ahola T, Kääriäinen L. Salonen A, et al. Curr Top Microbiol Immunol. 2005;285:139-73. doi: 10.1007/3-540-26764-6_5. Curr Top Microbiol Immunol. 2005. PMID: 15609503 Free PMC article. Review. - Alphavirus polymerase and RNA replication.
Pietilä MK, Hellström K, Ahola T. Pietilä MK, et al. Virus Res. 2017 Apr 15;234:44-57. doi: 10.1016/j.virusres.2017.01.007. Epub 2017 Jan 16. Virus Res. 2017. PMID: 28104453 Review.
Cited by
- Entry receptors - the gateway to alphavirus infection.
Zimmerman O, Holmes AC, Kafai NM, Adams LJ, Diamond MS. Zimmerman O, et al. J Clin Invest. 2023 Jan 17;133(2):e165307. doi: 10.1172/JCI165307. J Clin Invest. 2023. PMID: 36647825 Free PMC article. Review. - Activity, Template Preference, and Compatibility of Components of RNA Replicase of Eastern Equine Encephalitis Virus.
Lello LS, Miilimäe A, Cherkashchenko L, Omler A, Skilton R, Ireland R, Ulaeto D, Merits A. Lello LS, et al. J Virol. 2023 Jan 31;97(1):e0136822. doi: 10.1128/jvi.01368-22. Epub 2022 Dec 19. J Virol. 2023. PMID: 36533950 Free PMC article. - Inhibitors of Venezuelan Equine Encephalitis Virus Identified Based on Host Interaction Partners of Viral Non-Structural Protein 3.
Bakovic A, Bhalla N, Alem F, Campbell C, Zhou W, Narayanan A. Bakovic A, et al. Viruses. 2021 Aug 3;13(8):1533. doi: 10.3390/v13081533. Viruses. 2021. PMID: 34452398 Free PMC article. - Alphavirus-Induced Membrane Rearrangements during Replication, Assembly, and Budding.
Elmasri Z, Nasal BL, Jose J. Elmasri Z, et al. Pathogens. 2021 Aug 4;10(8):984. doi: 10.3390/pathogens10080984. Pathogens. 2021. PMID: 34451448 Free PMC article. Review. - nsP4 Is a Major Determinant of Alphavirus Replicase Activity and Template Selectivity.
Lello LS, Bartholomeeusen K, Wang S, Coppens S, Fragkoudis R, Alphey L, Ariën KK, Merits A, Utt A. Lello LS, et al. J Virol. 2021 Sep 27;95(20):e0035521. doi: 10.1128/JVI.00355-21. Epub 2021 Jul 28. J Virol. 2021. PMID: 34319783 Free PMC article.
References
- Arai, R., M. Geffard, and A. Calas. 1992. Intensification of labelings of the immunogold silver staining method by gold toning. Brain Res. Bull. 28:343-345. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources