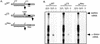In vivo association of the stability control protein alphaCP with actively translating mRNAs - PubMed (original) (raw)
In vivo association of the stability control protein alphaCP with actively translating mRNAs
Xinjun Ji et al. Mol Cell Biol. 2003 Feb.
Abstract
Posttranscriptional controls play a major role in eucaryotic gene expression. These controls are mediated by sequence-specific interactions of cis-acting determinants in target mRNAs with one or more protein factors. The positioning of a subset of these mRNA-protein (RNP) complexes within the 3' untranslated region (3' UTR) may allow them to remain associated with the mRNA during active translation. Robust expression of human alpha-globin mRNA during erythroid differentiation has been linked to formation of a binary complex between a KH-domain protein, alphaCP, and a 3' UTR C-rich motif. Detection of this "alpha-complex" has been limited to in vitro studies, and the functional state of the alpha-globin mRNA targeted by alphaCP has not been defined. In the present study we demonstrate that a significant fraction of alphaCP is associated with polysomal mRNA. Targeted analysis of the polysomal RNP complexes revealed that alphaCP is specifically bound to actively translating alpha-globin mRNA. The bound alphaCP is restricted to the poly(C)-rich 3' UTR motif and is dislodged when ribosomes are allowed to enter this region. These data validate the general importance of the 3' UTR as a sheltered site for RNP complexes and support a specific model in which the stabilizing function of alphaCP is mediated on actively translating target mRNAs.
Figures
FIG. 1.
hα-globin mRNA localizes to the polysome fraction of K562 cells. A clarified (S20) cytoplasmic extract from log-growth K562 was pelleted through a 30% sucrose cushion to separate prepolysome supernatant (S130) and polysomal (P) pellet fractions. hα-globin and GAPDH mRNAs were detected by RPA. The positions of the protected bands corresponding to GAPDH and α-globin are noted. The band above α-globin corresponds in size to the primary α-globin transcript and may be detecting contaminant genomic DNA. A 25-nt DNA ladder is shown (M).
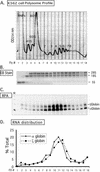
FIG. 2.
Distribution of hα- and ζ-globin mRNAs across a K562 cell polysome gradient. (A) Sucrose gradient profile of K562 cytosolic extract. The absorbance profile (OD254) of the gradient is shown. The top of the gradient is to the left; the positions of absorbance peaks corresponding to preribosomal RNPs, 40S, 60S, and 80S, and polysomes (2-, 3-, 4-, 5-, 6-, 7-, and 8-somes) are indicated. The 18 fractions (Fx #) collected for subsequent analysis are identified below the tracing. (B) Agarose gel electrophoresis of RNA extracted from the polysome gradient fractions. A 2-μg RNA sample from each fraction (in panel A) was applied to the gel and electrophoresed, and the abundant 28S, 18S, and 5S rRNAs were directly visualized by ethidium bromide staining. The distributions of these RNAs were consistent with the OD peak assignment (in panel A). (C) Detection of globin mRNAs across the K562 polysome gradient. Each gradient fraction was assessed for hα-globin and hζ-globin mRNAs by RPA with corresponding 32P-labeled probes. hα-globin and hζ-globin mRNAs protected probe fragments of 132 and 150 bp, respectively. (D) Distribution of globin mRNAs across the K562 polysome gradient. The contents of hα-globin and ζ-globin mRNAs in each fraction (in panel C) were quantified by PhosphorImager analysis. The amount of each mRNA species in each fraction is depicted as a percentage (ordinate) of the total for the corresponding species across the gradient.
FIG. 3.
αCP proteins are ribosome associated. A clarified K562 cell cytoplasmic extract (S20) was fractionated into prepolysomal (S130) and polysomal (P) fractions by sedimentation through a 30% sucrose cushion. Each of the three preparations was resolved on an SDS-PAGE gel; equal aliquots of S20 and S130 fractions and fivefold-concentrated aliquots of the polysome fraction were separated by SDS-PAGE. αCP2/KL and ribosomal L7a proteins were detected by Western blotting with corresponding antisera. The band at 37 kDa represents an isoform of αCP2 (αCP2KL). The positions of the molecular mass markers (not shown) are indicated on the left.
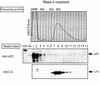
FIG. 4.
Characterization of the αCP-polysome interaction. Polysome aliquots (as in Fig. 3) were treated in the indicated manners and analyzed on 10-to-50% sucrose gradients. (A) No additional treatment. The OD254 is indicated (upper panel; Polysome Profile). Proteins in each fraction were precipitated, separated by SDS-12% PAGE, transferred to membranes, and probed with the indicated antibodies (Western blots). See the legend to Fig. 3 for details. (B) EDTA treatment. The polysome sample was resuspended in 20 mM EDTA (final concentration) prior to sucrose gradient fractionation. This treatment dissociates polysomes into 40S and 60S ribosome subunits. The splitting of the αCP signal seen in this Western blot is occasionally observed. (C) Treatment with 0.5 M KCl. The polysome fraction was brought to 0.5 M KCl (final concentration) prior to sucrose gradient fractionation. This treatment removes proteins loosely associated with the polysomes. (D) Treatment with 0.8 M KCl. The polysome fraction was supplemented with 0.8 M KCl (final concentration) prior to sucrose gradient fractionation. This treatment removes almost all proteins from the polysomes that are not intrinsic ribosomal proteins.
FIG. 5.
Association of αCP with polysomes is RNA dependent. The polysome fraction was prepared as detailed in the legend for Fig. 3, and an aliquot was treated with RNase A prior to sucrose gradient fractionation. Details of the analysis are as in the legend to Fig. 4.
FIG. 6.
In vivo association of αCP with polysome-bound hα-globin mRNA. Equal amounts of the prepolysomal (S130) and polysomal (P) fractions of K562 cytoplasmic extracts were individually immunoprecipitated with an antibody specific to the αCP2 and αCP2-KL isoforms. RNA extracted from the immunoprecipitates was assessed for specific mRNA content by RPA. This study was carried out independently four times with consistent results. (A) RPA using probes to hα-globin and GAPDH mRNAs. (B) RPA using probes to hα-globin and γ-globin mRNAs.
FIG. 7.
Interaction of αCP2 with α-globin mRNA is uniquely dependent on the 3′ UTR poly(C)-rich region and is sensitive to displacement by an antiterminated 80S. (A) Expression of hα-globin mRNAs with distinct 3′ UTRs (WT, Neu, and CS) in MEL/tTA cells. The structures of the three encoded mRNAs under the control of a tet promoter are shown. The positions of the translation start site (AUG), termination site (UAA), αCP binding site (protected region [PR]), and the antitermination mutation in the αCS (UAA → CAA) are shown. The cross-hatched box represents the substitution of a neutral sequence for the PR motif. (B) αCP binding on hα-globin mRNA is restricted to the C-rich 3′ UTR stability motif. Each of the indicated α-globin mRNAs was expressed in transfected cells from a corresponding plasmid. After 24 h of induction of expression in TET-deficient medium, the cells were lysed and the clarified cytoplasmic (S20) extracts were layered onto a 30% sucrose cushion. The isolated prepolysomal (S130) and polysomal (P) fractions were separately immunoprecipitated with antibody to αCP2 and αCP2-KL or with preimmune serum (PI). RNA was extracted from the starting material and from each immunoprecipitate. hα-globin and GAPDH mRNAs were detected by RPA. The origin of each sample is indicated above its respective lane. The positions of the RPA probes are indicated to the right of the gel. (C) Selective dissociation of αCP from the antiterminated αCS mRNA. MEL/tTA cells were separately transfected with pTet-WT and pTet-CS plasmids. The transfected genes were transcriptionally induced for 24 h in TET-deficient medium. TET was then added back to the medium at a concentration of 500 ng/ml for an additional 2 h. The cells were subsequently lysed and clarified (S10), and RNP complexes were immunoprecipitated with anti-αCP2 sera. mRNA content in the precipitate was analyzed by RPA as described for panel B. These studies were carried out independently three times with consistent results.
FIG. 7.
Interaction of αCP2 with α-globin mRNA is uniquely dependent on the 3′ UTR poly(C)-rich region and is sensitive to displacement by an antiterminated 80S. (A) Expression of hα-globin mRNAs with distinct 3′ UTRs (WT, Neu, and CS) in MEL/tTA cells. The structures of the three encoded mRNAs under the control of a tet promoter are shown. The positions of the translation start site (AUG), termination site (UAA), αCP binding site (protected region [PR]), and the antitermination mutation in the αCS (UAA → CAA) are shown. The cross-hatched box represents the substitution of a neutral sequence for the PR motif. (B) αCP binding on hα-globin mRNA is restricted to the C-rich 3′ UTR stability motif. Each of the indicated α-globin mRNAs was expressed in transfected cells from a corresponding plasmid. After 24 h of induction of expression in TET-deficient medium, the cells were lysed and the clarified cytoplasmic (S20) extracts were layered onto a 30% sucrose cushion. The isolated prepolysomal (S130) and polysomal (P) fractions were separately immunoprecipitated with antibody to αCP2 and αCP2-KL or with preimmune serum (PI). RNA was extracted from the starting material and from each immunoprecipitate. hα-globin and GAPDH mRNAs were detected by RPA. The origin of each sample is indicated above its respective lane. The positions of the RPA probes are indicated to the right of the gel. (C) Selective dissociation of αCP from the antiterminated αCS mRNA. MEL/tTA cells were separately transfected with pTet-WT and pTet-CS plasmids. The transfected genes were transcriptionally induced for 24 h in TET-deficient medium. TET was then added back to the medium at a concentration of 500 ng/ml for an additional 2 h. The cells were subsequently lysed and clarified (S10), and RNP complexes were immunoprecipitated with anti-αCP2 sera. mRNA content in the precipitate was analyzed by RPA as described for panel B. These studies were carried out independently three times with consistent results.
Similar articles
- Assembly of the alpha-globin mRNA stability complex reflects binary interaction between the pyrimidine-rich 3' untranslated region determinant and poly(C) binding protein alphaCP.
Chkheidze AN, Lyakhov DL, Makeyev AV, Morales J, Kong J, Liebhaber SA. Chkheidze AN, et al. Mol Cell Biol. 1999 Jul;19(7):4572-81. doi: 10.1128/MCB.19.7.4572. Mol Cell Biol. 1999. PMID: 10373506 Free PMC article. - Shared stabilization functions of pyrimidine-rich determinants in the erythroid 15-lipoxygenase and alpha-globin mRNAs.
Kong J, Sumaroka M, Eastmond DL, Liebhaber SA. Kong J, et al. Mol Cell Biol. 2006 Aug;26(15):5603-14. doi: 10.1128/MCB.01845-05. Mol Cell Biol. 2006. PMID: 16847316 Free PMC article. - An RNA-protein complex links enhanced nuclear 3' processing with cytoplasmic mRNA stabilization.
Ji X, Kong J, Liebhaber SA. Ji X, et al. EMBO J. 2011 May 27;30(13):2622-33. doi: 10.1038/emboj.2011.171. EMBO J. 2011. PMID: 21623344 Free PMC article. - Regulation of alpha-globin mRNA stability.
Waggoner SA, Liebhaber SA. Waggoner SA, et al. Exp Biol Med (Maywood). 2003 Apr;228(4):387-95. doi: 10.1177/153537020322800409. Exp Biol Med (Maywood). 2003. PMID: 12671183 Review. - An erythroid-enriched endoribonuclease (ErEN) involved in alpha-globin mRNA turnover.
Liu H, Kiledjian M. Liu H, et al. Protein Pept Lett. 2007;14(2):131-6. doi: 10.2174/092986607779816168. Protein Pept Lett. 2007. PMID: 17305599 Review.
Cited by
- RNA-binding protein PCBP2 modulates glioma growth by regulating FHL3.
Han W, Xin Z, Zhao Z, Bao W, Lin X, Yin B, Zhao J, Yuan J, Qiang B, Peng X. Han W, et al. J Clin Invest. 2013 May;123(5):2103-18. doi: 10.1172/JCI61820. Epub 2013 Apr 15. J Clin Invest. 2013. PMID: 23585479 Free PMC article. - The RNA-binding protein TcUBP1 up-regulates an RNA regulon for a cell surface-associated Trypanosoma cruzi glycoprotein and promotes parasite infectivity.
Sabalette KB, Romaniuk MA, Noé G, Cassola A, Campo VA, De Gaudenzi JG. Sabalette KB, et al. J Biol Chem. 2019 Jun 28;294(26):10349-10364. doi: 10.1074/jbc.RA118.007123. Epub 2019 May 21. J Biol Chem. 2019. PMID: 31113862 Free PMC article. - Specific enrichment of the RNA-binding proteins PCBP1 and PCBP2 in chief cells of the murine gastric mucosa.
Ghanem LR, Chatterji P, Liebhaber SA. Ghanem LR, et al. Gene Expr Patterns. 2014 Mar;14(2):78-87. doi: 10.1016/j.gep.2014.01.004. Epub 2014 Jan 28. Gene Expr Patterns. 2014. PMID: 24480778 Free PMC article. - PolyC-binding proteins enhance expression of the CDK2 cell cycle regulatory protein via alternative splicing.
Ji X, Humenik J, Yang D, Liebhaber SA. Ji X, et al. Nucleic Acids Res. 2018 Feb 28;46(4):2030-2044. doi: 10.1093/nar/gkx1255. Nucleic Acids Res. 2018. PMID: 29253178 Free PMC article. - The poly(rC)-binding protein alphaCP2 is a noncanonical factor in X. laevis cytoplasmic polyadenylation.
Vishnu MR, Sumaroka M, Klein PS, Liebhaber SA. Vishnu MR, et al. RNA. 2011 May;17(5):944-56. doi: 10.1261/rna.2587411. Epub 2011 Mar 28. RNA. 2011. PMID: 21444632 Free PMC article.
References
- Ashley, C. T., K. D. Wilkinson, D. Reines, and S. T. Warren. 1993. FMR1 protein: conserved RNP family domains and selective RNA binding. Science 262:563-566. - PubMed
- Atwater, J. A., R. Wisdom, and I. M. Verma. 1990. Regulated mRNA stability. Annu. Rev. Genet. 24:519-541. - PubMed
- Bernstein, P. L., D. J. Herrick, R. D. Prokipcak, and J. Ross. 1992. Control of c-myc mRNA half-life in vitro by a protein capable of binding to a coding region stability determinant. Genes Dev. 6:642-654. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 HL065449/HL/NHLBI NIH HHS/United States
- P01 CA072765/CA/NCI NIH HHS/United States
- CA72765/CA/NCI NIH HHS/United States
- HL 65449/HL/NHLBI NIH HHS/United States
- R37 HL065449/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources