A Bni4-Glc7 phosphatase complex that recruits chitin synthase to the site of bud emergence - PubMed (original) (raw)
A Bni4-Glc7 phosphatase complex that recruits chitin synthase to the site of bud emergence
Lukasz Kozubowski et al. Mol Biol Cell. 2003 Jan.
Abstract
Bni4 is a scaffold protein in the yeast Saccharomyces cerevisiae that tethers chitin synthase III to the bud neck by interacting with septin neck filaments and with Chs4, a regulatory subunit of chitin synthase III. We show herein that Bni4 is also a limiting determinant for the targeting of the type 1 serine/threonine phosphatase (Glc7) to the bud neck. Yeast cells containing a Bni4 variant that fails to associate with Glc7 fail to tether Chs4 to the neck, due in part to the failure of Bni4(V831A/F833A) to localize properly. Conversely, the Glc7-129 mutant protein fails to bind Bni4 properly and glc7-129 mutants exhibit reduced levels of Bni4 at the bud neck. Bni4 is phosphorylated in a cell cycle-dependent manner and Bni4(V831A/F833A) is both hyperphosphorylated and mislocalized in vivo. Yeast cells lacking the protein kinase Hsl1 exhibit increased levels of Bni4-GFP at the bud neck. GFP-Chs4 does not accumulate at the incipient bud site in either a bni4::TRP1 or a bni4(V831A/F833A) mutant but does mobilize to the neck at cytokinesis. Together, these results indicate that the formation of the Bni4-Glc7 complex is required for localization to the site of bud emergence and for subsequent targeting of chitin synthase.
Figures
Figure 1
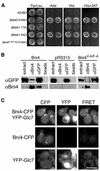
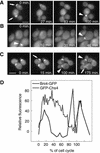
Interaction between Bni4 and Glc7. (A) Two-hybrid analysis of Bni4 and Glc7. Strain PJ69-4A was cotransformed with pHH149, encoding Glc7 fused to the Gal4 DNA-binding domain, and either pAR5 (BNI4 aa 70–892), pAR14 (BNI4 aa 1–779), pAR16 (BNI4 aa 1–892), or pAR19 (bni4 V-A/F-A aa 70–892), which encode the indicated Bni4 variants. Two representative transformants were plated on synthetic medium lacking tryptophane and leucine (SC-Trp/Leu) to select for the two plasmids. The two-hybrid interaction was assayed by replica plating the transformants onto SC-Ade, SC-His, or SC-His supplemented with 10 mM 3-amino-triazole (−His + 3AT). (B) Coimmunoprecipitation of Bni4 and Glc7. Strain KT1927 (GFP-GLC7, bni4::TRP1) was transformed with p366 (BNI4), the empty vector pRS316, and pAR17 (bni4 V-A/F-A). Immunoprecipitates were subjected to PAGE and immunoblots were probed using anti-GFP antibody (αGFP) or anti-Bni4 antibody (αBni4). (C) Strains YLK29 (Bni4-CFP, YFP-Glc7), YLK27 (Bni4-CFP) and YAB122 (YFP-Glc7) were imaged with CFP, YFP, and FRET filter sets. Bar, 5 μm.
Figure 2
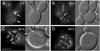
Localization of GFP-Glc7 to the bud neck is dependent on BNI4. (A) Montage of images taken at 20-min intervals from time-lapse experiments of diploid cells homozygous for BNI4 (strain KT1925 × KT1926) or bni4::TRP1 (strain KT1921 × KT1922). Arrows designate the location of _BNI4_-dependent GFP-Glc7 at the bud neck and the lack of GFP-Glc7 at the neck in bin4Δ cells. Arrowheads designate the accumulation of _BNI4_-independent GFP-Glc7 at the bud neck during cytokinesis. (B) Images of GFP-Glc7 (strain YAB608) in cells transformed with empty vector YEp351, low-copy Bni4-expressing vector (p366) or high copy Bni4-expressing vector (p365). (C) Fluorescence levels of GFP-Glc7 were quantified in the cytoplasm, at the bud necks and in the nuclei of the cells described in B. Levels for the cytoplasm were measured in the bud. Fluorescence levels in the nucleus were measured away from the nucleolus, which contains the highest levels of Glc7. Fluorescence levels in at least 20 cells from each transformed strain were quantified. A video supplement to Figure 2A was prepared from images taken at 10-min intervals. Bars (A and B), 5 μm.
Figure 3
Overexpression of GST-Bni4 causes elongated bud morphology and mislocalization of septins. Cells containing CDC10-GFP (YLK66) and expressing GST-Bni4 under galactose-inducible promoter (pLK4) were incubated in galactose medium at 37°C. Cells were imaged for Cdc10-GFP fluorescence at the designated times after the shift to galactose. After 2 h of incubation, Cdc10-GFP appeared in the form of patches at random locations on the cell membrane (A, arrowhead). Cdc10-GFP at the bud neck was also partially disorganized (A, arrow). At 5 h, CDC10-GFP appeared in short arced filaments dispersed in the cell membrane in addition to patches (B, arrow). Cdc10-GFP was miss-ing from bud necks. After 24 h of incubation, CDC10-GFP formed rings of ∼1 μm in diameter. Similar results were observed after incubation at 24 and 30°C. Bar, 5 μm.
Figure 4
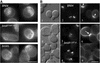
Effect of the bni4 V-A/F-A mutation on chitin synthesis at the bud neck. (A) Calcofluor staining in wild-type BNI4 (YLK74), bni4 V-A/F-A (YLK76), and bni4Δ1::TRP1 (YLK78) strains. Bud scars were of uniform size and shape in BNI4 strains whereas bud scars of bni4 V-A/F-A and bni4Δ1::TRP1 mutants were enlarged and irregular. (B) Colocalization of the septin Cdc10-GFP and chitin in BNI4 (YLK112) and bni4 V-A/F-A (YLK110) strains. In the BNI4 strain, relatively high levels of chitin were present at the bud neck throughout the cell cycle (3, arrowheads), whereas in bni4 V-A/F-A cells, higher levels of chitin at the bud neck were detected only in some large-budded cells (6, arrow). Bars, 5 μm.
Figure 5
Localization of GFP-Chs4 and Bni4-GFP by time-lapse imaging. (A) In the BNI4 cells (KT1972 cotransformed with pAR26 and p366), GFP-Chs4 accumulates at the site of the presumptive bud (0 min, top arrow) and it disappears soon after bud emergence. GFP-Chs4 reappears at the neck before cytokinesis (100 arrowhead). (B) In the bni4 V-A/F-A cells (KT1972 cotransformed with pAR26 and pAR17), GFP-Chs4 does not accumulate at the site of bud emergence. However, it appears at the bud neck before cytokinesis (0 and 46 min, arrowheads). (C) Localization of Bni4-GFP by time-lapse imaging (YLK80). Bni4-GFP appears at the future bud emergence site ∼15 min before budding (15 min, arrow) and stays at the bud neck at relatively high levels for 60–80% of the cell cycle (100 and 175 min, arrowheads). Close to cytokinesis, Bni4 forms a double ring that is hardly detectable (175 min, arrow). (D) Relative fluorescence of GFP-Chs4 (solid line) and Bni4-GFP (dotted line) was plotted as a function of the progress through the cell cycle (percentage of the cell cycle completed) by using the first appearance of GFP at the incipient bud site as the start point and the appearance of GFP at the incipient bud site in the mother cells in the next generation as the endpoint. Shown are representative data from three cells of each strain for which complete cell cycle data were obtained. A video supplement to Figure 5, A–C, was prepared from images taken at 5-min intervals. Bars (A and C), 5 μm.
Figure 6
Bni4V-A/F-A fails to localize properly to the bud neck. (A) Indirect immunofluorescence of Bni4 in a bni4Δ1::TRP1 strain (YLK78), a wild-type strain (YLK74), and in a bni4 V-A/F-A strain (YLK76), as described in Materials and Methods. Bni4 was visualized using anti-Bni4 antibody (DeMarini et al., 1997). Bar, 5 μm. (B) Immunoblot analysis of the strains described in A. (C) GFP fluorescence in bni4 V-A/F-A :GFP (YLK84) and BNI4:GFP (YLK80) strains.
Figure 7
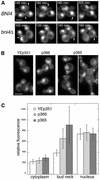
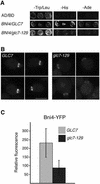
Levels of Bni4-YFP at the bud neck are reduced in glc7-129 cells. (A) Two-hybrid analysis of Bni4 and Glc7. Strain PJ69-4A was cotransformed with pAR5, encoding Bni4, and either pHH149 or pKT1703, which encode Glc7 and glc7-129, respectively. (B) Bni4-YFP was visualized in either KT2153 (GLC7) or in KT2155 (glc7-129) cells. Bar, 5 μm. (C) Fluorescence at the bud neck in the small- and medium-budded cells of GLC7 or glc7-129 strains from B was measured (p < 0.001).
Figure 8
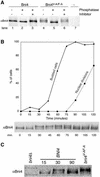
Bni4 is covalently modified. (A) Bni4 is phosphorylated and Bni4V-A/F-A is hyperphosphorylated. Immunoprecipitation with anti-Bni4 antibody (α-Bni4) was performed on lysates from cells expressing either Bni4 (lanes 1–3), Bni4V-A/F-A (lanes 4–6), or cells disrupted for BNI4 (lane 7) (strains YLK74, YLK76, and YLK78, respectively). Precipitates were MOCK-treated (lanes 1 and 4), treated with calf intestinal phosphatase (lanes 2 and 5), or treated with the phosphatase and the vanadate inhibitor (lanes 3 and 6). Samples were electrophoresed, blotted to membrane, and probed with α-Bni4 antibody. (B) Phosphorylation of Bni4 is cell cycle-dependent. Cells (DLY222) were arrested in G1 by incubation in medium containing sucrose. The arrest was released by exposing the cells to galactose. Samples were collected every 15 min and examined for budding (solid line), nuclear division (dotted line), and Bni4 mobility on PAGE (immunoblot), as detailed in the text. (C) Electrophoretic mobility of Bni4 in the samples collected at 30 min is similar to that of Bni4V-A/F-A from an asynchronous culture. Immunoblot with anti-Bni4 was done following SDS-PAGE separation of 15, 30, and 90 min. samples along with extract from the strain YLK76.
Figure 9
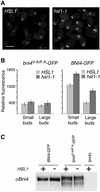
Effect of hsl1-1 deletion on the levels of Bni4-GFP and Bni4V-A/F-A-GFP at the bud neck. (A) Bni4-GFP in HSL1 (left, YLK45) and in hsl1-1 (right, YLK160) cells. Bar, 5 μm. (B) Relative fluorescence was measured for Bni4V-A/F-A-GFP (left) and Bni4-GFP (right) in HSL1 (YLK84, YLK45) and hsl1-1 (YLK162, YLK160) cells. Small-budded and large-budded cells were compared. For Bni4-GFP the difference between HSL1 and hsl1-1 in small- and large-budded cells was significant (p < 0.001). (C) Immunoblot analysis of extracts from Bni4-GFP and Bni4V-A/F-A-GFP in HSL1 and hsl1-1 strains. The immunoblots were probed with antibodies to Bni4.
Similar articles
- Changes in Bni4 localization induced by cell stress in Saccharomyces cerevisiae.
Larson JR, Kozubowski L, Tatchell K. Larson JR, et al. J Cell Sci. 2010 Apr 1;123(Pt 7):1050-9. doi: 10.1242/jcs.066258. Epub 2010 Mar 2. J Cell Sci. 2010. PMID: 20197406 Free PMC article. - Protein phosphatase type 1 directs chitin synthesis at the bud neck in Saccharomyces cerevisiae.
Larson JR, Bharucha JP, Ceaser S, Salamon J, Richardson CJ, Rivera SM, Tatchell K. Larson JR, et al. Mol Biol Cell. 2008 Jul;19(7):3040-51. doi: 10.1091/mbc.e08-02-0130. Epub 2008 May 14. Mol Biol Cell. 2008. PMID: 18480405 Free PMC article. - A septin-based hierarchy of proteins required for localized deposition of chitin in the Saccharomyces cerevisiae cell wall.
DeMarini DJ, Adams AE, Fares H, De Virgilio C, Valle G, Chuang JS, Pringle JR. DeMarini DJ, et al. J Cell Biol. 1997 Oct 6;139(1):75-93. doi: 10.1083/jcb.139.1.75. J Cell Biol. 1997. PMID: 9314530 Free PMC article. - Septin-associated protein kinase Gin4 affects localization and phosphorylation of Chs4, the regulatory subunit of the Baker's yeast chitin synthase III complex.
Gohlke S, Heine D, Schmitz HP, Merzendorfer H. Gohlke S, et al. Fungal Genet Biol. 2018 Aug;117:11-20. doi: 10.1016/j.fgb.2018.05.002. Epub 2018 May 12. Fungal Genet Biol. 2018. PMID: 29763674 - Regulation of cell polarity through phosphorylation of Bni4 by Pho85 G1 cyclin-dependent kinases in Saccharomyces cerevisiae.
Zou J, Friesen H, Larson J, Huang D, Cox M, Tatchell K, Andrews B. Zou J, et al. Mol Biol Cell. 2009 Jul;20(14):3239-50. doi: 10.1091/mbc.e08-12-1255. Epub 2009 May 20. Mol Biol Cell. 2009. PMID: 19458192 Free PMC article.
Cited by
- Substrates of the MAPK Slt2: Shaping Yeast Cell Integrity.
González-Rubio G, Sastre-Vergara L, Molina M, Martín H, Fernández-Acero T. González-Rubio G, et al. J Fungi (Basel). 2022 Apr 4;8(4):368. doi: 10.3390/jof8040368. J Fungi (Basel). 2022. PMID: 35448599 Free PMC article. Review. - Changes in Bni4 localization induced by cell stress in Saccharomyces cerevisiae.
Larson JR, Kozubowski L, Tatchell K. Larson JR, et al. J Cell Sci. 2010 Apr 1;123(Pt 7):1050-9. doi: 10.1242/jcs.066258. Epub 2010 Mar 2. J Cell Sci. 2010. PMID: 20197406 Free PMC article. - Ypi1, a positive regulator of nuclear protein phosphatase type 1 activity in Saccharomyces cerevisiae.
Bharucha JP, Larson JR, Gao L, Daves LK, Tatchell K. Bharucha JP, et al. Mol Biol Cell. 2008 Mar;19(3):1032-45. doi: 10.1091/mbc.e07-05-0499. Epub 2008 Jan 2. Mol Biol Cell. 2008. PMID: 18172024 Free PMC article. - Regulation of expression, activity and localization of fungal chitin synthases.
Rogg LE, Fortwendel JR, Juvvadi PR, Steinbach WJ. Rogg LE, et al. Med Mycol. 2012 Jan;50(1):2-17. doi: 10.3109/13693786.2011.577104. Epub 2011 Apr 28. Med Mycol. 2012. PMID: 21526913 Free PMC article. Review. - Long-Chain Polyprenols Promote Spore Wall Formation in Saccharomyces cerevisiae.
Hoffmann R, Grabińska K, Guan Z, Sessa WC, Neiman AM. Hoffmann R, et al. Genetics. 2017 Dec;207(4):1371-1386. doi: 10.1534/genetics.117.300322. Epub 2017 Oct 4. Genetics. 2017. PMID: 28978675 Free PMC article.
References
- Beullens M, Van Eynde A, Vulsteke V, Connor J, Shenolikar S, Stalmans W, Bollen M. Molecular determinants of nuclear protein phosphatase-1 regulation by NIPP-1. J Biol Chem. 1999;274:14053–14061. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases