Sam68 RNA binding protein is an in vivo substrate for protein arginine N-methyltransferase 1 - PubMed (original) (raw)
Sam68 RNA binding protein is an in vivo substrate for protein arginine N-methyltransferase 1
Jocelyn Côté et al. Mol Biol Cell. 2003 Jan.
Abstract
RNA binding proteins often contain multiple arginine glycine repeats, a sequence that is frequently methylated by protein arginine methyltransferases. The role of this posttranslational modification in the life cycle of RNA binding proteins is not well understood. Herein, we report that Sam68, a heteronuclear ribonucleoprotein K homology domain containing RNA binding protein, associates with and is methylated in vivo by the protein arginine N-methyltransferase 1 (PRMT1). Sam68 contains asymmetrical dimethylarginines near its proline motif P3 as assessed by using a novel asymmetrical dimethylarginine-specific antibody and mass spectrometry. Deletion of the methylation sites and the use of methylase inhibitors resulted in Sam68 accumulation in the cytoplasm. Sam68 was also detected in the cytoplasm of PRMT1-deficient embryonic stem cells. Although the cellular function of Sam68 is unknown, it has been shown to export unspliced human immunodeficiency virus RNAs. Cells treated with methylase inhibitors prevented the ability of Sam68 to export unspliced human immunodeficiency virus RNAs. Other K homology domain RNA binding proteins, including SLM-1, SLM-2, QKI-5, GRP33, and heteronuclear ribonucleoprotein K were also methylated in vivo. These findings demonstrate that RNA binding proteins are in vivo substrates for PRMT1, and their methylation is essential for their proper localization and function.
Figures
Figure 1
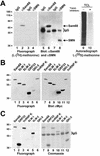
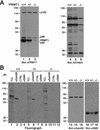
In vivo methylation of Sam68 and other KH domain RNA binding proteins. (A) In vivo methylation labeling was performed by metabolically labeling cells with [
l
-[methyl-3H]methionine for 3 h in the presence of cycloheximide and chloramphenicol. The cell lysates were immunoprecipitated with anti-Sam68 (mAb 7-1) and anti-SMN antibodies. The 3H-labeled proteins were separated by SDS-PAGE and visualized by fluorography (lane 1–4; exp. 24 h), and the proteins were visualized by immunoblotting with anti-Sam68 and anti-SMN antibodies (lanes 5–8). The migration of Sam68 and SMN is shown. The heavy chain of IgG is indicated with a bracket. TCLs of HeLa cells metabolically labeled with
l
-[35S]methionine in the absence (lane 9; −) or presence (lane 10; +) of protein synthesis inhibitors were separated by SDS-PAGE and visualized by autoradiography (exp. 24 h). (B) Metabolic labeling for the in vivo methylation was performed 24 h after transfection of myc-epitope–tagged Sam68, SLM-1, SLM-2, GRP33, QKI-5, and hnRNP K expression vectors in HeLa cells. The cell lysates were immunoprecipitated with anti-myc antibodies and the bound 3H-labeled proteins were separated by SDS-PAGE and visualized either by fluorography (lanes 1–6; exp. 12 h) or by immunoblotting with the anti-myc antibody (lanes 7–12). (C) Endogenous protein methyltransferase activity coimmunoprecipitates with KH domain-containing proteins. HeLa cells were transfected with myc-epitope–tagged Sam68, SLM-1, SLM-2, GRP33, QKI-5, and hnRNP K. The cells were lyzed and myc immunoprecipitations were incubated with [3H-_methyl_]_S_-adenosyl-
l
-methionine in vitro. The immunoprecipitated proteins were visualized by Coomassie staining (lanes 7–12) or by fluorography (lanes 1–6; exp. 6 h). The migration of the heavy chain of IgG is indicated.
Figure 2
Association of Sam68 and other KH domain RNA binding proteins with PRMT1. (A) Purified recombinant GST or GST-PRMT1 were separated by SDS-PAGE and immunoblotted with anti-PRMT1 antibodies (lanes 1 and 2). The band at ∼70 kDa represents GST-PRMT1. HeLa cell extracts were separated by SDS-PAGE and immunoblotted with NRS, anti-PRMT1 antibodies, and anti-PRMT1 immunoabsorbed with the antigenic peptide. The migration of the four immunogenic proteins (p120, p48, p45, and p42) recognized by anti-PRMT1 antibodies is indicated on the right (lanes 3–5). The predominant band of ∼45 kDa most likely represents PRMT1 as predicted. The migration of the molecular mass markers in kilodaltons is shown on the left. (B) Myc-tagged Sam68, SLM-1, SLM-2, GRP33, hnRNP K, and QKI-5 were expressed in HeLa and immunoprecipitated with control IgG or anti-myc antibodies. Aliquots of TCLs (10% input) and the immunoprecipitated proteins were resolved by SDS-PAGE and immunoblotted with anti-PRMT1 antibodies. The migration of the PRMT1 immunogenic proteins and the heavy chain of IgG are indicated. Longer exposure of lanes 1–12 also reveals p42 and p48 (our unpublished data). (C) Equivalent expression of myc-epitope–tagged KH domain RNA proteins was confirmed by immunoblotting an aliquot of TCLs.
Figure 3
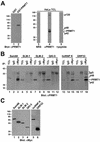
Sam68 is asymmetrically dimethylated in vivo as detected by using novel anti-aDMA-specific antibodies. (A) Generation of an anti-aDMA antibody. Peptide (24aDMA; see B for sequence) was used as an antigenic peptide to inject rabbits. The specificity of the purified polyclonal antibody (ASYM24) was examined by ELISA with a control unrelated peptide (AD1 peptide), unmethylated “backbone” peptide 24, or an identical peptide containing symmetrical dimethylated arginines (24sDMA). The quantity of peptide bound to the plate was plotted against the immunoreactivity of the ASYM24 antibody. Each point represents the average of three separate experiments. (B) Specificity of the antibody was examined by using a variety of unrelated aDMA containing peptides with neighboring proline-rich sequences. One microgram of the indicated peptides was coated per ELISA well. WASP P1 and P2 denote the Wiscott-Aldrich Syndrome protein proline motifs 1 and 2 containing aDMA. hnRNPK P1 and P2 denote hnRNPK proline motifs 1 and 2 containing aDMA. Sam68 P0, P3, and P4 denote the Src substrate activated in mitosis of 68-kDa proline motifs 0, 3, and 4 containing aDMA. The unmethylated P3 peptide was also used as control (Sam68 P3). Each bar denotes greater than n > 8 from three separate experiments. (C) Myc-tagged Sam68 was transfected in HeLa cells and immunoprecipitated with control (IgG) or anti-Myc antibodies. Aliquots of TCLs (10% input) and the bound proteins were separated by SDS-PAGE and visualized by immunoblotting with anti-myc, NRS, ASYM24, and ASYM24 antibodies absorbed with the antigenic peptide (+24 pept.). The migration of Sam68, the heavy chain of IgG, and endogenous proteins (p110 and p75) is indicated. (D) GST-Sam68 fusion protein was either left untreated (−) or incubated with GST-PRMT1 under in vitro methylation conditions in the presence of “cold” _S_-adenosyl-
l
-methionine. An aliquot of each reaction was resolved by SDS-PAGE, transferred to nitrocellulose membranes, and immunoblotted with anti-Sam68 AD1 antibodies (lanes 1 and 2) and ASYM24 (lanes 3 and 4). The molecular mass markers are shown on the left in kilodaltons. (E) HeLa cells were cotransfected with expression vectors encoding myc-Sam68 and myc-PRMT1. The cells were lysed in sample buffer and the proteins separated by SDS-PAGE and immunoblotted with either anti-myc or ASYM24 antibodies. The migration of myc-PRMT1, myc-Sam68, p110, and p75 is indicated. (F) HeLa cells or HEK293 (293) cells were lysed and immunoprecipitated with control antibodies (NRS and mouse IgG) and anti-Sam68 antibodies (pAb s.c.333, pAb AD1, mAb 7-1). The proteins were separated by SDS-PAGE and immunoblotted with ASYM24. The migration of Sam68, the heavy chain of IgG, p110, and p75 is shown on the right. The molecular mass markers are shown on the left.
Figure 4
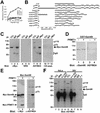
Sam68 RG-rich sequences are methylated in vivo and regulate protein localization. (A) Schematic diagram representing the various Sam68 proteins is shown. The position of the KH domain and the neighboring N-terminal of KH (NK) and C-terminal of KH (CK) are indicated. The GSG domain encompasses the NK, KH, and CK domains. The gray boxes represent regions where RG repeats are present in Sam68. The C-terminal nuclear localization signal (NLS) of Sam68 is represented by a stippled box. (B) In vivo methylation labeling was performed 24 h after DNA transfection of the expression vectors encoding the proteins depicted in A. The cell lysates were immunoprecipitated with anti-myc antibodies and the 3H-labeled proteins were separated by SDS-PAGE and visualized by fluorography (lanes 1–4; exp. 12 h) and by immunoblotting by using anti-myc antibodies (lanes 5–8). (C) HeLa cells plated on glass coverslips were transfected with myc-epitope–tagged Sam68 (1 and 5), the Sam68 deletion protein that is no longer methylated (SΔN:Δ280–339; 2 and 6), a slightly longer deleted mutant that is methylated (SΔN:Δ294–339; 3 and 7), and a Sam68 missing the C terminus, including the NLS (SΔN:ΔC; 4 and 8). Twenty-four hours after transfection, the cells were fixed and immunostained by using anti-myc antibodies followed by a secondary antibody conjugated to Alexa 546 (red; 1–4). Nuclei were visualized using the DNA-specific stain 4,6-diamidino-2-phenylindole (blue; 5–8).(D) HeLa cells plated on glass coverslips were transfected with myc-epitope–tagged PRMT1 followed by indirect immunofluorescence by using anti-myc epitope-tagged antibodies followed by a secondary antibody conjugated to Alexa 546 (red). Nuclei were visualized using the DNA specific stain 4,6-diamidino-2-phenylindole.
Figure 5
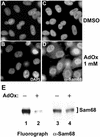
Sam68 accumulates in the cytoplasm in the presence of methylase inhibitors. HeLa cells were treated with either DMSO (A and C) or with the methyltransferase inhibitor AdOx at 1 mM (B and D) for 24 h. The cells were fixed and immunostained with the anti-Sam68 AD1 antibody followed by a secondary antibody conjugated to Alexa 488 (green; C and D). Nuclei were visualized by the DNA-specific stain 4,6-diamidino-2-phenylindole (blue; A and B). (E) HeLa cells were incubated with AdOx for 24 h preceding an in vivo methylation labeling. Immunoprecipitations were performed with anti-Sam68 AD1 antibodies. Samples were separated by 10% SDS-PAGE, transferred to a polyvinylidene difluoride membrane, sprayed with Enhance, and exposed to film overnight. The 3H-labeled proteins were visualized by fluorography (left), and the same membrane was immunoblotted with anti-Sam68 AD1 antibodies (right).
Figure 6
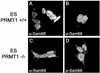
In vivo methylation of Sam68 is abrogated in prmt1 −/− ES cells. (A) Wild-type ES cells (+/+) as well as ES cells heterozygotes (+/−) or homozygotes (−/−) for a null mutation in the prmt1 gene were harvested in lysis buffer and subjected to immunoblotting by using our anti-PRMT1 and ASYM24 antibodies. Molecular weight markers (in kilodaltons) are shown on the left of each panel. (B) In vivo methylation labeling was performed by metabolically labeling cells with
l
-[_methyl_-3H]methionine for 3 h in the presence of cycloheximide and chloramphenicol. The cell lysates were immunoprecipitated with anti-Sam68 (AD1), normal rabbit serum (CTRL), or anti-SMN antibodies. The 3H-labeled proteins in TCLs (10% input) and in immunoprecipitates were separated by SDS-PAGE and visualized by fluorography (lane 1–12; exp. 24 h). Immunoblotting was performed on total cell lysates by using anti-Sam68 (lanes 13–15) and anti-SMN (lanes 16–18) antibodies to confirm equal levels of proteins.
Figure 7
Sam68 accumulates in the cytoplasm of PRMT1 −/− ES cells. Wild-type ES cells (PRMT1 +/+; A and B) or ES cells homozygotes (PRMT1 −/−; C and D) for a null mutation in the prmt1 gene were fixed and immunostained using the anti-Sam68 AD1 antibody followed by a secondary antibody conjugated to Alexa 488. Cells were then visualized using a confocal microscope (Carl Zeiss).
Figure 8
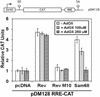
Methylation is necessary for the ability of Sam68 to function in the nuclear export of HIV RNAs. A schematic diagram of the Rev responsive element reporter CAT construct is shown. The splice acceptor and donor sites are indicated as SA and SD. CAT indicates the chloramphenicol acetyltransferase cDNA and RRE is the HIV Rev response element. COS7 cells were transfected with the RRE-CAT reporter plasmid pDM128 in the presence of either an empty plasmid (pcDNA), Rev, an inactive mutant of Rev (Rev M10), or Sam68. After transfection, cells were treated with increasing concentration of AdOx or with the vehicle alone (DMSO; −AdOx). CAT activity was normalized to β-galactosidase activity. Each bar represents CAT activity from at least three separate experiments.
Similar articles
- Arginine methylation of Sam68 and SLM proteins negatively regulates their poly(U) RNA binding activity.
Rho J, Choi S, Jung CR, Im DS. Rho J, et al. Arch Biochem Biophys. 2007 Oct 1;466(1):49-57. doi: 10.1016/j.abb.2007.07.017. Epub 2007 Aug 6. Arch Biochem Biophys. 2007. PMID: 17764653 - hCAF1, a new regulator of PRMT1-dependent arginine methylation.
Robin-Lespinasse Y, Sentis S, Kolytcheff C, Rostan MC, Corbo L, Le Romancer M. Robin-Lespinasse Y, et al. J Cell Sci. 2007 Feb 15;120(Pt 4):638-47. doi: 10.1242/jcs.03357. Epub 2007 Jan 30. J Cell Sci. 2007. PMID: 17264152 - Protein arginine methylation of SERBP1 by protein arginine methyltransferase 1 affects cytoplasmic/nuclear distribution.
Lee YJ, Hsieh WY, Chen LY, Li C. Lee YJ, et al. J Cell Biochem. 2012 Aug;113(8):2721-8. doi: 10.1002/jcb.24151. J Cell Biochem. 2012. PMID: 22442049 - Reaching for the stars: Linking RNA binding proteins to diseases.
Richard S. Richard S. Adv Exp Med Biol. 2010;693:142-57. Adv Exp Med Biol. 2010. PMID: 21189691 Review. - Emerging roles for Sam68 in adipogenesis and neuronal development.
Vogel G, Richard S. Vogel G, et al. RNA Biol. 2012 Sep;9(9):1129-33. doi: 10.4161/rna.21409. Epub 2012 Sep 1. RNA Biol. 2012. PMID: 23018781 Free PMC article. Review.
Cited by
- Protein arginine methyltransferase 3-induced metabolic reprogramming is a vulnerable target of pancreatic cancer.
Hsu MC, Tsai YL, Lin CH, Pan MR, Shan YS, Cheng TY, Cheng SH, Chen LT, Hung WC. Hsu MC, et al. J Hematol Oncol. 2019 Jul 19;12(1):79. doi: 10.1186/s13045-019-0769-7. J Hematol Oncol. 2019. PMID: 31324208 Free PMC article. - Nck adapter proteins: functional versatility in T cells.
Lettau M, Pieper J, Janssen O. Lettau M, et al. Cell Commun Signal. 2009 Feb 2;7:1. doi: 10.1186/1478-811X-7-1. Cell Commun Signal. 2009. PMID: 19187548 Free PMC article. - Protein arginine methylation in viral infection and antiviral immunity.
Zheng K, Chen S, Ren Z, Wang Y. Zheng K, et al. Int J Biol Sci. 2023 Oct 24;19(16):5292-5318. doi: 10.7150/ijbs.89498. eCollection 2023. Int J Biol Sci. 2023. PMID: 37928266 Free PMC article. Review. - Arginine methylation enhances the RNA chaperone activity of the West Nile virus host factor AUF1 p45.
Friedrich S, Schmidt T, Schierhorn A, Lilie H, Szczepankiewicz G, Bergs S, Liebert UG, Golbik RP, Behrens SE. Friedrich S, et al. RNA. 2016 Oct;22(10):1574-91. doi: 10.1261/rna.055269.115. Epub 2016 Aug 12. RNA. 2016. PMID: 27520967 Free PMC article. - Transcriptional repression of hypoxia-inducible factor-1 (HIF-1) by the protein arginine methyltransferase PRMT1.
Lafleur VN, Richard S, Richard DE. Lafleur VN, et al. Mol Biol Cell. 2014 Mar;25(6):925-35. doi: 10.1091/mbc.E13-07-0423. Epub 2014 Jan 22. Mol Biol Cell. 2014. PMID: 24451260 Free PMC article.
References
- Baldwin GS, Carnegie PR. Specific enzymic methylation of an arginine in the experimental allergic encephalomyelitis protein from human myelin. Science. 1971;171:579–581. - PubMed
- Bedford MT, Frankel A, Yaffe MB, Clarke S, Leder P, Richard S. Arginine methylation inhibits the binding of proline-rich ligands to Src homology 3, but not WW, domains. J Biol Chem. 2000;275:16030–16036. - PubMed
- Brahms H, Raymackers J, Union A, de Keyser F, Meheus L, Luhrmann R. The C-terminal RG dipeptide repeats of the spliceosomal Sm proteins D1 and D3 contain symmetrical dimethylarginines, which form a major B-cell epitope for anti-Sm autoantibodies. J Biol Chem. 2000;275:17122–17129. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases