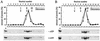The VirB4 family of proposed traffic nucleoside triphosphatases: common motifs in plasmid RP4 TrbE are essential for conjugation and phage adsorption - PubMed (original) (raw)
The VirB4 family of proposed traffic nucleoside triphosphatases: common motifs in plasmid RP4 TrbE are essential for conjugation and phage adsorption
Christian Rabel et al. J Bacteriol. 2003 Feb.
Abstract
Proteins of the VirB4 family are encoded by conjugative plasmids and by type IV secretion systems, which specify macromolecule export machineries related to conjugation systems. The central feature of VirB4 proteins is a nucleotide binding site. In this study, we asked whether members of the VirB4 protein family have similarities in their primary structures and whether these proteins hydrolyze nucleotides. A multiple-sequence alignment of 19 members of the VirB4 protein family revealed striking overall similarities. We defined four common motifs and one conserved domain. One member of this protein family, TrbE of plasmid RP4, was genetically characterized by site-directed mutagenesis. Most mutations in trbE resulted in complete loss of its activities, which eliminated pilus production, propagation of plasmid-specific phages, and DNA transfer ability in Escherichia coli. Biochemical studies of a soluble derivative of RP4 TrbE and of the full-length homologous protein R388 TrwK revealed that the purified forms of these members of the VirB4 protein family do not hydrolyze ATP or GTP and behave as monomers in solution.
Figures
FIG. 1.
Purification of RP4 TrbEHNΔ2 (A) and R388 TrwK (B). Aliquots of extracts and pooled peak fractions (Table 4) were resolved with SDS-15% polyacrylamide gels and stained with Coomassie blue. Cells were lysed either by SDS or by Brij 58-lysozyme; the prominent band at an apparent mass of 14 kDa in panel A, lane d, is lysozyme. (A) Lane a, molecular mass standard; lanes b and c, SDS extracts of noninduced and IPTG-induced cells, respectively; lane d, fraction I (amount of total protein loaded onto the gel, 56.2 μg); lane e, fraction II (9.9 μg); lane f, fraction III (9.2 μg); lane g, fraction IV (10.1 μg). (B) Lane a, molecular mass standard; lanes b and c, SDS extracts of noninduced and IPTG-induced cells, respectively; lane d, fraction I (amount of total protein loaded onto the gel, 13.2 μg); lane e, fraction II (7.1 μg); lane f, fraction III (8.2 μg); lane g, fraction IV (7.4 μg).
FIG. 2.
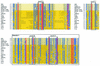
Conserved motifs in proteins belonging to the VirB4 family. Walker boxes A (motif A) and B (motif B), motifs C and D, and domain I are conserved in a multiple-sequence alignment (red, conserved residues; blue, residues conserved in more than 70% of the sequences; gray, residues with similar side chains). The subfamily of TrbE-like proteins is indicated by a light yellow background, and the subfamily of VirB4-like proteins is indicated by a dark yellow background. Amino acids in RP4 TrbE which were subjected to site-directed mutagenesis are indicated by solid diamonds above the sequences. With the exception of TrbE and VirB4 encoded by pTiA6, only the name of the plasmid or bacterium encoding the protein homologous to VirB4 is given. The following VirB4 homologs were examined (the numbers in parentheses are accession numbers): RP4, TrbE (AAA26431); R751, TrbE (NP_044243); pNGR234a, TrbEb (AAB92432); pRi1724, TrbE (BAB16246); pTiA6 TrbE (AAB95097); pTiA6 VirB4 (AAF77164); Brucella melitensis VirB4 (B.m.) (AAL53269); pSB102, TraE (CAC79179); Bordetella pertussis PtlC (B.p.) (B47301); R388, TrwK (CAC78982); pXF51, XFa0007 (AAF85576); Bartonella henselae VirB4 (B.h.) (AAF00942); pVT745, MagB03 (AAG24434); R6K, Pilx4 (CAC20141); pKM101, TraB (I79267); Rickettsia prowazekii VirB4 (R.p.) (NP_220495); Legionella pneumophila LvhB4 (L.p.) (CAB60053); R721, TraE (NP_065362); and H. pylori HP0544 (H.p.) (NP_207340).
FIG. 3.
Amino acid substitutions in RP4 TrbE and common motifs in the VirB4 protein family. Protein RP4 TrbE is represented by the gray bar. The predicted TMH and motifs A to D are represented by solid boxes. Each of the mutations influenced the Pil, Dps, and Tra phenotypes (Table 2). Proposed consensus sequences of the common motifs found in proteins belonging to the VirB4 family are shown below the bar. aa, amino acid; NBS, nucleotide binding site.
FIG. 4.
Phenotypes of RP4 TrbE deletion mutants. Wild-type TrbE and N- and C-terminal deletions are represented by gray bars. The predicted TMH and motifs A to D (Fig. 2) are indicated by solid boxes only in wild-type TrbE (TrbE wt). The His6 tag at the N terminus of TrbEHNΔ2 is indicated by the dark gray box. The phenotypes of the mutants are indicated on the right. aa, amino acids; Overexpr., overexpression; n.d., not determined.
FIG. 5.
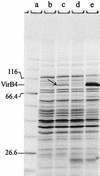
Overproduction of pTiA6 VirB4 in E. coli strain BL21. Heterologous overexpression of virB4 in E. coli is inhibited by different codon usage. The virB4 gene contains 11 codons for arginine, nine codons for leucine, and two codons for proline, which are rare in E. coli. Plasmid pACYC-RIL encodes the tRNAs argU (AGA, AGG), ileY (AUA), and leuW (CUA). Freshly transformed strains BL21(pCR6) and BL21(pCR6, pACYC-RIL) were induced with 1 mM IPTG and grown for 5 h. Crude cell extracts of induced and noninduced cells were separated on an SDS-15% polyacrylamide gel and stained with Coomassie blue. Lane a, molecular mass standard; lane b, noninduced BL21(pCR6); lane c, induced BL21(pCR6); lane d, noninduced BL21(pCR6, pACYC-RIL); lane e, induced BL21(pCR6, pACYC-RIL). The arrow in lane c indicates the faint band of VirB4 resulting from induction in the absence of pACYC-RIL; the prominent band in lane e was shown to be VirB4.
FIG. 6.
Glycerol density gradient centrifugation of RP4 TrbEHNΔ2 (A) and R388 TrwK (B). Purified proteins TrbEHNΔ2 (fraction IV, 120 μl, 0.2 mg) (Table 4) and TrwK (fraction IV, 120 μl, 0.2 mg) (Table 4) were laid on a 3.8-ml, linear, 15 to 35% (wt/vol) glycerol gradient in buffer E. Protein samples were prepared in buffer E. For analysis of conformational changes, the samples were mixed with 1 mM ATP and 10 mM MgCl2 and incubated for 10 min at 30°C prior to centrifugation. Centrifugation was carried out at 272,000 × g (rmax) for 18 h at 4°C. Fifteen fractions, each containing 15 drops of the gradient, were collected. Fifty microliters of each fraction and a 10-μl aliquot (lane a) of TrbEHNΔ2 or TrwK were separated on SDS-15% polyacrylamide gels. The gels were stained with Coomassie blue and scanned with a Personal densitometer (Molecular Dynamics). The amount of protein in each lane was quantified by using the software ImageQuant 5.0 (Molecular Dynamics). The relative amount of protein in each fraction is shown in the graphs (▪, ATP and MgCl2 added; □, ATP and MgCl2 omitted). The arrows indicate the peak positions of the reference proteins (arrow I, catalase [240 kDa, _s_20,w = 11.3]; arrow II, aldolase [158 kDa, _s_20,w = 7.8]; arrow III, BSA [66 kDa, _s_20,w = 4.4]).
Similar articles
- ATPase activity and oligomeric state of TrwK, the VirB4 homologue of the plasmid R388 type IV secretion system.
Arechaga I, Peña A, Zunzunegui S, del Carmen Fernández-Alonso M, Rivas G, de la Cruz F. Arechaga I, et al. J Bacteriol. 2008 Aug;190(15):5472-9. doi: 10.1128/JB.00321-08. Epub 2008 Jun 6. J Bacteriol. 2008. PMID: 18539740 Free PMC article. - Promiscuous DNA transfer system of Agrobacterium tumefaciens: role of the virB operon in sex pilus assembly and synthesis.
Kado CI. Kado CI. Mol Microbiol. 1994 Apr;12(1):17-22. doi: 10.1111/j.1365-2958.1994.tb00990.x. Mol Microbiol. 1994. PMID: 7914664 Review. - Analysis of the sequence and gene products of the transfer region of the F sex factor.
Frost LS, Ippen-Ihler K, Skurray RA. Frost LS, et al. Microbiol Rev. 1994 Jun;58(2):162-210. doi: 10.1128/mr.58.2.162-210.1994. Microbiol Rev. 1994. PMID: 7915817 Free PMC article. Review.
Cited by
- Structure of the VirB4 ATPase, alone and bound to the core complex of a type IV secretion system.
Walldén K, Williams R, Yan J, Lian PW, Wang L, Thalassinos K, Orlova EV, Waksman G. Walldén K, et al. Proc Natl Acad Sci U S A. 2012 Jul 10;109(28):11348-53. doi: 10.1073/pnas.1201428109. Epub 2012 Jun 27. Proc Natl Acad Sci U S A. 2012. PMID: 22745169 Free PMC article. - Persistence of a pKPN3-like CTX-M-15-encoding IncFIIK plasmid in a Klebsiella pneumonia ST17 host during two years of intestinal colonization.
Löhr IH, Hülter N, Bernhoff E, Johnsen PJ, Sundsfjord A, Naseer U. Löhr IH, et al. PLoS One. 2015 Mar 4;10(3):e0116516. doi: 10.1371/journal.pone.0116516. eCollection 2015. PLoS One. 2015. PMID: 25738592 Free PMC article. - Conjugative coupling proteins interact with cognate and heterologous VirB10-like proteins while exhibiting specificity for cognate relaxosomes.
Llosa M, Zunzunegui S, de la Cruz F. Llosa M, et al. Proc Natl Acad Sci U S A. 2003 Sep 2;100(18):10465-70. doi: 10.1073/pnas.1830264100. Epub 2003 Aug 18. Proc Natl Acad Sci U S A. 2003. PMID: 12925737 Free PMC article. - Autoinhibitory regulation of TrwK, an essential VirB4 ATPase in type IV secretion systems.
Peña A, Ripoll-Rozada J, Zunzunegui S, Cabezón E, de la Cruz F, Arechaga I. Peña A, et al. J Biol Chem. 2011 May 13;286(19):17376-82. doi: 10.1074/jbc.M110.208942. Epub 2011 Mar 24. J Biol Chem. 2011. PMID: 21454654 Free PMC article. - Biochemical Analysis of CagE: A VirB4 Homologue of Helicobacter pylori Cag-T4SS.
Shariq M, Kumar N, Kumari R, Kumar A, Subbarao N, Mukhopadhyay G. Shariq M, et al. PLoS One. 2015 Nov 13;10(11):e0142606. doi: 10.1371/journal.pone.0142606. eCollection 2015. PLoS One. 2015. PMID: 26565397 Free PMC article.
References
- Bamford, D. H., L. Rouhiainen, K. Takkinen, and H. Soderlund. 1981. Comparison of the lipid-containing bacteriophages PRD1, PR3, PR4, PR5 and L17. J. Gen. Virol. 57:365-373. - PubMed
- Baron, C., D. O'Callaghan, and E. Lanka. 2002. Bacterial secrets of secretion: EuroConference on the Biology of Type IV Secretion Processes. Mol. Microbiol. 43:1359-1365. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources