Apollo: a sequence annotation editor - PubMed (original) (raw)
Review
. 2002;3(12):RESEARCH0082.
doi: 10.1186/gb-2002-3-12-research0082. Epub 2002 Dec 23.
S M J Searle, N Harris, M Gibson, V Lyer, J Richter, C Wiel, L Bayraktaroglu, E Birney, M A Crosby, J S Kaminker, B B Matthews, S E Prochnik, C D Smithy, J L Tupy, G M Rubin, S Misra, C J Mungall, M E Clamp
Affiliations
- PMID: 12537571
- PMCID: PMC151184
- DOI: 10.1186/gb-2002-3-12-research0082
Review
Apollo: a sequence annotation editor
S E Lewis et al. Genome Biol. 2002.
Abstract
The well-established inaccuracy of purely computational methods for annotating genome sequences necessitates an interactive tool to allow biological experts to refine these approximations by viewing and independently evaluating the data supporting each annotation. Apollo was developed to meet this need, enabling curators to inspect genome annotations closely and edit them. FlyBase biologists successfully used Apollo to annotate the Drosophila melanogaster genome and it is increasingly being used as a starting point for the development of customized annotation editing tools for other genome projects.
Figures
Figure 1
A standard view in Apollo, showing a 250 kb segment of Drosophila chromosome arm 2L. The background colors for the result (gray), annotation (light blue) and sequence (white) panels are configurable. The features and annotations on the forward strand are shown above the white sequence panel, and those on the reverse strand are shown below it. There are seven tiers (or rows) of data presented here in the results panel (the number of tiers is configurable). On the forward strand from top to bottom these results are promoter predictions (yellow), P-element insertions (turquoise triangles), peptide homologies to other species (orange), peptide homologies to Drosophila (red), Drosophila EST alignments (light green), Drosophila mRNA alignments (bright and dark green) and gene prediction results (lavender). The genomic region navigation bar is above the data displays and the navigational and zoom controls for the current genomic region in the display are below. The user may increase the genomic region visible in the display by using the 'Expand' button. They may also move to adjacent regions by clicking on the '<' (5' upstream) and '>' (3' downstream) buttons or move to entirely new regions by changing the chromosome arm and the start and end positions. Within the display the most basic movement operations (zooming and scrolling) are available from the controls at the bottom of the display.
Figure 2
Some of the feature appearance options that are available. Each feature's shape, visibility, color and panel position is configurable. The result panel can display the complete portfolio of computational results. The top three rows of this figure use small vertical green lines to indicate the position of start codons in the genomic sequence in all three possible translation frames. The second set of three rows does the same for stop codons (red). This enables curators to easily discern ORFs. Beneath this, in orange, are BLASTX hits to other species. Every row represents a separate alignment in this expanded view (see Figure 3 for a discussion of the expanded view). In each alignment the high-scoring pairs (HSP) are separated by two parallel lines to indicate alignment gaps. Seeing what features share edges is important during annotation for adjusting exon-intron boundaries. When an item is selected, corresponding edges of other items that end at the same nucleotide are highlighted with a white line (arrow A). The small turquoise triangle indicates the site of a P-element insertion. The rows of light green rectangles are Sim4 alignments of Drosophila ESTs to the genome. When a 5' and a 3' EST are derived from the same cDNA clone we connect them with a dashed line (arrow B). This contrasts with the solid lines used to represent the gaps, corresponding to introns, introduced into an EST to permit its alignment to the genome. The final piece of computational evidence shown in this figure is the gene prediction, shown in lavender. In this example, we have intentionally represented introns in two different ways - as straight or peaked lines - as an illustration of Apollo's configurable graphics. In the annotation panel, the curator has created two alternative transcripts for this gene, each of which is supported by multiple pieces of EST evidence. Individual exons that the curator has selected are outlined in yellow. The translation start and stop sites are shown as green and red vertical lines, respectively. Each individual curator has a signature color; any annotation that this particular curator creates is shown in bright blue. In the sequence panel the scale is drawn in red to indicate that this gene is on the reverse strand.
Figure 3
A tier organizes a collection of one or more feature types into a single horizontal row in the view. One of the viewing options is whether the features within that tier are 'collapsed' or 'expanded'. In the collapsed view (top) features lie on top of one another and thus appear along a single line. In this view the EST alignments (shown in light green) are collapsed. In the expanded view (below) the tier disallows overlapping features and thus as many lines as needed are used. The individual EST alignments (one line per EST) now appear separately. The tiers that flank the EST tier, alignments of peptide from other species (orange) or from Drosophila (red), are shown collapsed in both the top and bottom panels.
Figure 4
This figure shows the feature description tables. Each row in the top panel describes a single feature. The columns provide the feature type (here designated as 'organism' name), the aligned accession, the genomic position of the alignment and the BLASTX score. Clicking on any column heading will sort the rows according to the values held in that column (the initial sorting order is specified in the configuration file). Detailed information for the selected feature in this table (highlighted in yellow) appears in a second table, shown in the lower panel. The table header displays the name of the aligned sequence, its full length and a full description. The columns are configurable, and for each feature type only those columns that have been specified for that type will be shown. In this example of BLASTX alignments to other species, the available information includes the BLASTX score, BLASTX expectation and the location and length of the alignment on both query and subject sequences.
Figure 5
A textual display provides detailed information for individual annotations. Annotations can be selected either from the menu on the left or by selecting an annotation in any other window. The types of annotations that can be generated by a curator are part of the initial configuration and appear in the menu shown above. A list of comments in a controlled vocabulary that can be used by the curator to add remarks about an annotation is also provided as part of the configuration (one example is shown above in the lower middle portion of the panel). These standardized comments facilitate future querying.
Figure 6
Zooming provides additional information. In the top panel, which shows an intermediate level of zoom, the start and stop codons in all three frames appear as green and red tick marks, then the BLASTX similarities to other species (orange bars) and finally BLASTX similarities to Drosophila peptides (red bars). When the level of zoom is increased as shown as the lower panel, the sequence (either nucleic acid or peptide) of any aligned sequences is loaded and displayed to enable curators to directly compare predicted peptides. An individual start or stop codon, such as the start codon shown selected in this example, can be dragged onto the annotation to explicitly specify translation start and end sites for protein coding transcripts in the curator's annotation (blue).
Figure 7
Apollo provides a variety of search capabilities, including moving to an absolute base pair position, locating an annotation or aligned sequence by name or accession, or finding an exact sequence match (above). The graphical interface centers the view over the selected result. The interface also makes it possible to bookmark locations (below). This allows curators to quickly return to areas they are actively working on.
Figure 8
An overview of the synteny between mouse and human is shown in Apollo's synteny display. The central (larger) chromosomes are the human chromosomes, which are flanked by their syntenic mouse chromosomes. Apollo clusters DNA-DNA matches into syntenic regions, eliminating any short paralogous DNA-DNA hits. Selecting a segment displays a menu showing the chromosomal coordinates of the match in the genome sequence of both organisms and offering the user two options. The first opens a gene-level synteny view, illustrated in Figure 9. The second option reads the raw DNA-DNA matches that were used to produce the synteny and displays a sequence-level view of the similarities between the two genomes.
Figure 9
A detailed synteny view in Apollo showing part of human chromosome 20 at the top and part of mouse chromosome 2 at the bottom is illustrated. In the middle are links between orthologous genes as identified by the Ensembl synteny-generating software. The links are crossed because the genes in the top panel and in the bottom panel are in different orientations on their respective chromosomes. Although this figure only shows gene links, other types of link may also be displayed. Information about the genes from both genomes are read from Ensembl databases and information about orthologous gene pairs can either be read from an Ensembl compara database or from a flat file. The central panel can be used to scroll back and forth along the syntenic region or to center the display on a particular region by clicking on one of the colored matches. The top and bottom Apollo panels behave just as a normal, single Apollo panel and can be zoomed, scrolled, collapsed and also link out to a web page (as configured by the user).
Figure 10
Fine control of editing requires close examination of the gene structure. This is accomplished in the exon editor panel, which provides a sequence-level view of gene models. When the exon editor is launched, the region displayed is highlighted in the main Apollo display (blue rectangle in lower panel), and this main display rectangle can be moved along the sequence to control the region shown in the exon editor. Using the contextual menu in the exon editor panel, changes can be made to introns and exons, as well as to precise splice junctions and translation starts. Annotations appear in the viewer with alternating colors for individual exons, and clicking on a transcript selects it and causes a graphical representation (or glyph) of the transcript to appear at the bottom of the window. This transcript glyph shows the translation start and stop sites, and can be displayed with or without introns. In the case illustrated, the intron-exon structure of the gene is shown; the numbers in the exons indicate which frame of the translated genomic sequence is utilized in that exon. Clicking on the different portions of the glyph allows one to quickly navigate to specific regions of the transcript. A sequence search query is also available, as is retrieval of genomic and translated sequence.
Figure 11
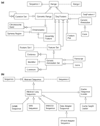
Categories of data models. The Apollo data models fall into one of two broad categories (or are a descriptive auxiliary class, for example, Comment): (a) a location on a sequence; or (b) a sequence. The corresponding Java superclasses are Range and AbstractSequence, respectively. The inheritance hierarchy from these two central classes is shown here, but some minor classes and relationships are omitted to simplify this description. Each class or interface is drawn as a rectangle, and interfaces have the suffix 'I' included in their class name (an interface specifies the methods that the class is required to implement). Lines ending with an open-headed arrow-point indicate the superclass and subclass relationships and a dotted line connects an interface to a class that implements that interface. Thus, both GenomicRange and SeqFeature are subclasses of the base class Range (which implements the Range I interface) and in turn FeatureSet, GenericAnnotation, FeaturePair, and AssemblyFeature are specializations of SeqFeature. Similarly an SRSSequence is a subclass of AbstractLazySequence, which in turn is a subclass AbstractSequence. Each of these subclasses inherits all the methods of their parent class and may extend the model's behavior either by adding new methods or by overriding the inherited methods. In addition to inheritance, the connecting lines also depict other types of relationships, with the terminus indicating the potential cardinality. Thus, a GenericAnnotationSet must have at least one, but may possibly have more, pieces of Evidence associated with it (drawn as a single crossbar to indicate 'at least one' and a triangular tripod to represent 'many'), one Identifier, which maintains synonyms and database cross references (a single crossbar indicates 'one and only one'), and may have zero or more optional Comments (drawn as a triangular tripod to represent 'many' and a single circle to indicate 'none'). Thus, a FeatureSet is both itself a SeqFeature that is composed of one or more component SeqFeatures. This enables Transcripts to be composed of a set of Exons, or an alignment to be composed of a set of high-scoring pairs. Likewise a CurationSet (across two different species) may contain component CurationSets (for the individual species) to enable comparative analysis.
Similar articles
- Using Apollo to browse and edit genome annotations.
Misra S, Harris N. Misra S, et al. Curr Protoc Bioinformatics. 2006 Jan;Chapter 9:9.5.1-9.5.28. doi: 10.1002/0471250953.bi0905s12. Curr Protoc Bioinformatics. 2006. PMID: 18428771 - Apollo2Go: a web service adapter for the Apollo genome viewer to enable distributed genome annotation.
Klee K, Ernst R, Spannagl M, Mayer KF. Klee K, et al. BMC Bioinformatics. 2007 Aug 30;8:320. doi: 10.1186/1471-2105-8-320. BMC Bioinformatics. 2007. PMID: 17760972 Free PMC article. - Apollo: Democratizing genome annotation.
Dunn NA, Unni DR, Diesh C, Munoz-Torres M, Harris NL, Yao E, Rasche H, Holmes IH, Elsik CG, Lewis SE. Dunn NA, et al. PLoS Comput Biol. 2019 Feb 6;15(2):e1006790. doi: 10.1371/journal.pcbi.1006790. eCollection 2019 Feb. PLoS Comput Biol. 2019. PMID: 30726205 Free PMC article. - An integrated computational pipeline and database to support whole-genome sequence annotation.
Mungall CJ, Misra S, Berman BP, Carlson J, Frise E, Harris N, Marshall B, Shu S, Kaminker JS, Prochnik SE, Smith CD, Smith E, Tupy JL, Wiel C, Rubin GM, Lewis SE. Mungall CJ, et al. Genome Biol. 2002;3(12):RESEARCH0081. doi: 10.1186/gb-2002-3-12-research0081. Epub 2002 Dec 23. Genome Biol. 2002. PMID: 12537570 Free PMC article. Review. - The Drosophila melanogaster genome sequencing and annotation projects: a status report.
Drysdale R. Drysdale R. Brief Funct Genomic Proteomic. 2003 Jul;2(2):128-34. doi: 10.1093/bfgp/2.2.128. Brief Funct Genomic Proteomic. 2003. PMID: 15239934 Review.
Cited by
- Assembly and comparative analysis of the complete mitochondrial genome of Fritillaria ussuriensis Maxim. (Liliales: Liliaceae), an endangered medicinal plant.
Xie P, Wu J, Lu M, Tian T, Wang D, Luo Z, Yang D, Li L, Yang X, Liu D, Cheng H, Tan J, Yang H, Zhu D. Xie P, et al. BMC Genomics. 2024 Aug 8;25(1):773. doi: 10.1186/s12864-024-10680-w. BMC Genomics. 2024. PMID: 39118028 Free PMC article. - Atlas - a data warehouse for integrative bioinformatics.
Shah SP, Huang Y, Xu T, Yuen MM, Ling J, Ouellette BF. Shah SP, et al. BMC Bioinformatics. 2005 Feb 21;6:34. doi: 10.1186/1471-2105-6-34. BMC Bioinformatics. 2005. PMID: 15723693 Free PMC article. - ParameciumDB in 2011: new tools and new data for functional and comparative genomics of the model ciliate Paramecium tetraurelia.
Arnaiz O, Sperling L. Arnaiz O, et al. Nucleic Acids Res. 2011 Jan;39(Database issue):D632-6. doi: 10.1093/nar/gkq918. Epub 2010 Oct 14. Nucleic Acids Res. 2011. PMID: 20952411 Free PMC article. - Munich information center for protein sequences plant genome resources: a framework for integrative and comparative analyses 1(W).
Schoof H, Spannagl M, Yang L, Ernst R, Gundlach H, Haase D, Haberer G, Mayer KF. Schoof H, et al. Plant Physiol. 2005 Jul;138(3):1301-9. doi: 10.1104/pp.104.059188. Plant Physiol. 2005. PMID: 16010004 Free PMC article. - Creating a honey bee consensus gene set.
Elsik CG, Mackey AJ, Reese JT, Milshina NV, Roos DS, Weinstock GM. Elsik CG, et al. Genome Biol. 2007;8(1):R13. doi: 10.1186/gb-2007-8-1-r13. Genome Biol. 2007. PMID: 17241472 Free PMC article.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
- Churchill GA. Stochastic models for heterogeneous DNA sequences. Bull Math Biol. 1989;51:79–94. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases