EpsinR: an AP1/clathrin interacting protein involved in vesicle trafficking - PubMed (original) (raw)
EpsinR: an AP1/clathrin interacting protein involved in vesicle trafficking
Ian G Mills et al. J Cell Biol. 2003.
Abstract
EpsinR is a clathrin-coated vesicle (CCV) enriched 70-kD protein that binds to phosphatidylinositol-4-phosphate, clathrin, and the gamma appendage domain of the adaptor protein complex 1 (AP1). In cells, its distribution overlaps with the perinuclear pool of clathrin and AP1 adaptors. Overexpression disrupts the CCV-dependent trafficking of cathepsin D from the trans-Golgi network to lysosomes and the incorporation of mannose-6-phosphate receptors into CCVs. These biochemical and cell biological data point to a role for epsinR in AP1/clathrin budding events in the cell, just as epsin1 is involved in the budding of AP2 CCVs. Furthermore, we show that two gamma appendage domains can simultaneously bind to epsinR with affinities of 0.7 and 45 microM, respectively. Thus, potentially, two AP1 complexes can bind to one epsinR. This high affinity binding allowed us to identify a consensus binding motif of the form DFxDF, which we also find in gamma-synergin and use to predict that an uncharacterized EF-hand-containing protein will be a new gamma binding partner.
Figures
Figure 1.
The ENTH domain of epsinR. (A) The ENTH domain of epsinR was modeled on epsin1 ENTH with Ins(1,4,5)P3 bound. The major difference in surface electrostatic potential (red − 10 kT e−1; blue + 10 kT e−1) is in the PtdInsP binding pocket where several positively charged residues are missing in epsinR ENTH domain. (B) Ribbon diagram of the modeled epsinR ENTH domain showing the residues in the binding pocket and the succession of hydrophobic residues on the outer surface of helix zero (α0), just like in epsin1 ENTH domain. (C) Sequence homologies between epsin1 homologues and epsinR homologues. Both Drosophila (D) and C. elegans (Ce) have one homologue of each, but humans have three homologues of epsin1 (1–3), and yeast (Sc, Saccharomyces cerevisiae) also has multiple homologues. Hydrophobic residues on the outer surface of helix zero are marked in yellow. Conserved (blue) and nonconserved (green) Ptd(4,5)P2 binding residues and other major differences (orange) are marked. Some of the key residues in the human epsinR sequence referred to in the paper are numbered. (D) Domain structure of epsin1 compared with epsinR. Clathrin binding motifs (pink), DPW/DLF adaptor binding motifs (purple) and regions of alternative splicing (dotted lines) are shown. The splice site at amino acid 460 is an insert of the residues QPLQNVSTVLQKPNPLYN. Below are shown all the constructs, mutations, truncations and peptides used in the paper. The ubiquitin-interacting motifs (UIMs) and the Eps15 binding motifs (NPFs) found in epsin1 are not present in epsinR. We also know that there are additional clathrin binding motifs in epsinR (at least one in the N2 domain) that we have not mapped.
Figure 2.
EpsinR binds to PtdIns(4)P and PtdIns(5)P in vitro. (A) Coomassie-stained gel of liposome-binding assays with epsinR ENTH domain, the PtdIns(4)P targeted GFP tagged PH domain of OSBP, and the PtdIns(3)P targeted PX domain of p40_phox_. Pellet (P) and supernatant (S). (B) PtdInsP strips confirm the lipid specificity of epsin1 ENTH domain for PtdIns(4,5)P2 and show that epsinR prefers PtdInsPs with a lower charge density. The R29L mutation weakens the interaction, and with the double mutation D34G + R67L, we lose the 4P and 5P interactions. OSBP also shows a very similar specificity to epsinR ENTH domain. The labeling on the first strip applies to the other panels.
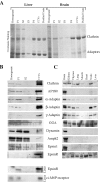
Figure 3.
EpsinR, a ubiquitous protein enriched in CCVs. (A) Purification of rat liver and brain CCVs. A stronger adaptor band is seen by Coomassie staining in the liver CCV preparation, although by electron microscopy, the liver CCVs are not as pure as the brain CCVs (not depicted). (B) Brain fractions were blotted for various CCV-enriched and -nonenriched proteins, and these were compared with epsin1 and epsinR in the same preparation. Blots for epsinR and the cation-independent M6P receptor are also shown for liver fractions. (C) Brain-specific distribution of epsin1 compared with the ubiquitous distribution of epsinR. Equal concentrations of rat tissue were loaded.
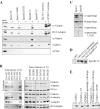
Figure 4.
Clathrin and AP1 adaptors bind to epsinR. (A) Epsin1 and epsinR constructs were tested for binding to adaptors, clathrin, and AP180 in rat brain extract. Epsin1, Eps15 and AP180 all bind to α- and β-adaptins but only epsinR binds to γ-adaptin (asterisk). All constructs were GST-tagged, and +/− indicates the presence/absence of brain extract. (B) Truncations of N1 narrowed the major clathrin/adaptor-binding domain to 291–426 (N3), and further truncations abolish clathrin and then adaptor binding (Coomassie-stained gel). GGAs also bind to the N3 construct, and the first deletion affects the interaction (blot). Point mutagenesis shows that D422R primarily affects clathrin binding over adaptor binding while AP2 adaptors are affected by the D349 mutation. AP1 adaptors are affected by D349 and D371 mutations. GGA binding does not follow the same pattern as clathrin or the multimeric adaptors. (C) Full-length Myc-epsin1 and Myc-epsinR expressed in COS cells bind to the GST-appendage domains of α-, β-, and γ-adaptin, but not the L762E γ-adaptin appendage. (D) GST constructs of GGA1 and GGA2 appendage domains and the N3 construct of epsinR bind to full-length epsinR in brain extract. (E) Clathrin assembly is not promoted by the N3 domain of epsinR at neutral pH. Controls with β-adaptin appendage + hinge domain and full-length AP180 both show different extents of assembly. Clathrin in pellet (P) and supernatant (S) fractions after spinning are shown.
Figure 5.
Two γ-adaptin appendage domains can bind simultaneously with high affinity to epsinR. (A) Affinity measurement by calorimetric titrations for α-, β-, and γ-appendage domains and GGA appendage domain with epsinR N3 constructs. Where the stoichiometry of the interaction was 2:1, the data showed a robust fit to a two-site model, and the KDs for both sites are shown. The bold lettering indicates the protein in the syringe. (B) Peptides from epsinR and γ-synergin binding to the γ-adaptin appendage domain. Profiles of typical calorimetric titrations and the integrated normalized data are shown on the right. A comprehensive table of calorimetric data is available in Table SI.
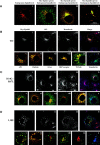
Figure 6.
Subcellular localization of epsinR. As a convention throughout the figure, myc-epsinR and mutants are labeled green and the endogenous proteins are labeled red. (A) Endogenous epsinR shows a perinuclear enrichment (Ai) with a very different distribution to Myc-epsin1 (green in Aii), but it colocalizes with overexpressed Myc-epsinR (green in Aiii). Colocalization is orange/yellow. We observed that the epsinR perinuclear compartment was frequently enlarged, and in extreme examples much of the overexpressed epsinR was accumulated there (Aiv). Endogenous epsinR is also accumulated in this compartment (Aiv). (B) Colocalization of Myc-epsinR with endogenous AP1 (ii and v), transferrin (iii and x), clathrin (vi), GGA (vii), cation-independent mannose-6-phosphate receptors (M6P, viii), and TGN46 (ix). Panels v–x are closer views of the perinuclear regions of cells stained for overexpressed epsinR and the indicated marker. There is much orange/yellow color in the perinuclear region of the cell implying a great deal of colocalization, but precise colocalization is hard to define in this region because of the accumulation of so many compartments. Transferrin (panel x) is labeled blue and colocalization with AP1 is cyan. (C and D) Colocalization of various markers with the D34G + R67L and the L10E mutants of epsinR, respectively. Labeling is the same as in B. Links to original images and extra data can be found at
http://www.jcb.org/cgi/content/full/jcb.200208023/DC1
.
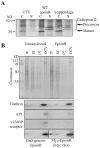
Figure 7.
Disruption of CCV trafficking from the TGN. (A) Overexpression of Myc-epsinR caused mistargeting of pro-cathepsin D. COS cells, either untransfected (UTC) or transfected with WT Myc-epsinR or Myc γ-appendage domain were pulse-chased with 35S, and both-cell associated (C) and -secreted (S) cathepsin D were immunoprecipitated and gels were phosphorimaged. The secreted forms are indicated by the asterisks. (B) Reduced incorporation of the cation-independent M6P receptor into purified CCVs in epsinR transfected COS cells.
Similar articles
- Binding of GGA2 to the lysosomal enzyme sorting motif of the mannose 6-phosphate receptor.
Zhu Y, Doray B, Poussu A, Lehto VP, Kornfeld S. Zhu Y, et al. Science. 2001 Jun 1;292(5522):1716-8. doi: 10.1126/science.1060896. Science. 2001. PMID: 11387476 - Clathrin adaptor epsinR is required for retrograde sorting on early endosomal membranes.
Saint-Pol A, Yélamos B, Amessou M, Mills IG, Dugast M, Tenza D, Schu P, Antony C, McMahon HT, Lamaze C, Johannes L. Saint-Pol A, et al. Dev Cell. 2004 Apr;6(4):525-38. doi: 10.1016/s1534-5807(04)00100-5. Dev Cell. 2004. PMID: 15068792 - A SNARE-adaptor interaction is a new mode of cargo recognition in clathrin-coated vesicles.
Miller SE, Collins BM, McCoy AJ, Robinson MS, Owen DJ. Miller SE, et al. Nature. 2007 Nov 22;450(7169):570-4. doi: 10.1038/nature06353. Nature. 2007. PMID: 18033301 - [Regulatory mechanisms of the clathrin adaptor molecules AP-1 and GGAs].
Kametaka S, Bonifacino JS. Kametaka S, et al. Tanpakushitsu Kakusan Koso. 2008 Dec;53(16 Suppl):2046-52. Tanpakushitsu Kakusan Koso. 2008. PMID: 21038583 Review. Japanese. No abstract available. - Adaptors for clathrin coats: structure and function.
Owen DJ, Collins BM, Evans PR. Owen DJ, et al. Annu Rev Cell Dev Biol. 2004;20:153-91. doi: 10.1146/annurev.cellbio.20.010403.104543. Annu Rev Cell Dev Biol. 2004. PMID: 15473838 Review.
Cited by
- EPSIN1 and MTV1 define functionally overlapping but molecularly distinct _trans_-Golgi network subdomains in Arabidopsis.
Heinze L, Freimuth N, Rößling AK, Hahnke R, Riebschläger S, Fröhlich A, Sampathkumar A, McFarlane HE, Sauer M. Heinze L, et al. Proc Natl Acad Sci U S A. 2020 Oct 13;117(41):25880-25889. doi: 10.1073/pnas.2004822117. Epub 2020 Sep 28. Proc Natl Acad Sci U S A. 2020. PMID: 32989160 Free PMC article. - Phagocytic receptor signaling regulates clathrin and epsin-mediated cytoskeletal remodeling during apoptotic cell engulfment in C. elegans.
Shen Q, He B, Lu N, Conradt B, Grant BD, Zhou Z. Shen Q, et al. Development. 2013 Aug;140(15):3230-43. doi: 10.1242/dev.093732. Development. 2013. PMID: 23861060 Free PMC article. - Genome-wide identification of Aedes albopictus long noncoding RNAs and their association with dengue and Zika virus infection.
Azlan A, Obeidat SM, Theva Das K, Yunus MA, Azzam G. Azlan A, et al. PLoS Negl Trop Dis. 2021 Jan 22;15(1):e0008351. doi: 10.1371/journal.pntd.0008351. eCollection 2021 Jan. PLoS Negl Trop Dis. 2021. PMID: 33481791 Free PMC article. - Drosophila liquid facets-Related encodes Golgi epsin and is an essential gene required for cell proliferation, growth, and patterning.
Lee JH, Overstreet E, Fitch E, Fleenor S, Fischer JA. Lee JH, et al. Dev Biol. 2009 Jul 1;331(1):1-13. doi: 10.1016/j.ydbio.2009.03.029. Epub 2009 Apr 17. Dev Biol. 2009. PMID: 19376106 Free PMC article. - The aftiphilin/p200/gamma-synergin complex.
Hirst J, Borner GH, Harbour M, Robinson MS. Hirst J, et al. Mol Biol Cell. 2005 May;16(5):2554-65. doi: 10.1091/mbc.e04-12-1077. Epub 2005 Mar 9. Mol Biol Cell. 2005. PMID: 15758025 Free PMC article.
References
- Balch, W.E., W.G. Dunphy, W.A. Braell, and J.E. Rothman. 1984. Reconstitution of the transport of protein between successive compartments of the Golgi measured by the coupled incorporation of N-acetylglucosamine. Cell. 39:405–416. - PubMed
- Boehm, M., and J.S. Bonifacino. 2002. Genetic analyses of adaptin function from yeast to mammals. Gene. 286:175–186. - PubMed
- Brett, T.J., L.M. Traub, and D.H. Fremont. 2002. Accessory protein recruitment motifs in clathrin-mediated endocytosis. Structure. 10:797–809. - PubMed
- Chen, H., S. Fre, V.I. Slepnev, M.R. Capua, K. Takei, M.H. Butler, P.P. Di Fiore, and P. De Camilli. 1998. Epsin is an EH-domain-binding protein implicated in clathrin-mediated endocytosis. Nature. 394:793–797. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials