PPARdelta is a very low-density lipoprotein sensor in macrophages - PubMed (original) (raw)
PPARdelta is a very low-density lipoprotein sensor in macrophages
Ajay Chawla et al. Proc Natl Acad Sci U S A. 2003.
Abstract
Although triglyceride-rich particles, such as very low-density lipoprotein (VLDL), contribute significantly to human atherogenesis, the molecular basis for lipoprotein-driven pathogenicity is poorly understood. We demonstrate that in macrophages, VLDL functions as a transcriptional regulator via the activation of the nuclear receptor peroxisome proliferator-activated receptor delta. The signaling components of native VLDL are its triglycerides, whose activity is enhanced by lipoprotein lipase. Generation of peroxisome proliferator-activated receptor delta null macrophages verifies the absolute requirement of this transcription factor in mediating the VLDL response. Thus, our data reveal a pathway through which dietary triglycerides and VLDL can directly regulate gene expression in atherosclerotic lesions.
Figures
Figure 1
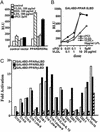
PPARδ is activated by the VLDL particle. (A) VLDL particle transactivates full-length PPARδ. Transient transfection experiments were performed in triplicate in CV1 cells using the AOx-PPRE3 luciferase reporter construct (0.1 μg) and the CMX-PPARδ/CMX-RXRα expression vectors (10 ng each) as described in Materials and Methods. Transfected cells were cultured in lipoprotein-deficient FBS and treated with ligands or lipoproteins for 24 h before collection for reporter gene analysis. Luciferase activity was normalized for transfection efficiency to an internal β-galactosidase control. (B) VLDL particle activates via the ligand binding domain of PPARδ. CV-1 cells were cotransfected with the chimeric receptor, GAL4DBD-PPARδLBD, and the UAS3 luciferase reporter gene. Transfected cells were subsequently treated with varying concentrations of cPGI, VLDL, or VLDL coincubated with 10 μg/ml LPL. (C) Triglycerides present in VLDL transactivate PPARδ. Transient transfections were performed in CV-1 cells with GAL-PPAR (α, δ, and γ) and UAS3 luciferase reporter. Triglycerides were coincubated with LPL (10 μg/ml) in 1% BSA before their addition to the medium (50 μM final concentration).
Figure 2
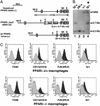
Generation of PPARδ-deficient ES cells and macrophages. (A) Targeting scheme for generation of PPARδ null ES cells. The G418-sensitive ES cell clone IIIA4 (23) consists of a mixture of cells heterozygous for the WT PPARδ allele (+, Bottom) and either a null “floxed-out” allele (−, Top) or a conditional “floxed” allele (ck, not shown). This clone was retargeted with a construct containing a lacZ gene knocked-in, which disrupts PPARδ upstream of the first zinc finger DNA-binding motif (lz, Middle). Patterned bars below each allele represent the corresponding predictions of _Eco_RI-flanked DNA fragments when probed with a 3′-external probe. Restriction sites: E, _Eco_RI; Nh, _Nhe_I. (B) Southern blot analysis of PPARδ null ES cells. The individual daughter clones generated by retargeting the IIIA4 cells contain different biallelic combinations of PPARδ: cells heterozygous for the WT and the newly introduced lacZ knock-in allele (+/lz; lane 1); two G418-resistant clones carrying the original IIIA4 allelic combinations ck/+ and +/− (lanes 2 and 3, respectively) and a likely nonhomologous integration of the neoR gene; and the PPARδ null ES clone analyzed functionally in this study (lz/−, lane 4; *), in which the lz allele integrated in place of the remaining WT allele of clone IIIA4. Allelic identity was further confirmed using a 5′ external probe (not shown). (C) FACS analysis of PPARδ+/+ and _PPARδ_−/− ES cell-derived macrophages. Macrophages were generated from ES cells as described in Materials and Methods. WT or PPARδ null macrophages were stained with the indicated antibodies (dark gray) or isotype control (light gray). Macrophage cell-surface markers include F4/80, Mac-1 (CD11b/CD18), and 2.4G2 (FcγRII/FcγRIII). Gr.1 (Ly-6G), a granulocyte lineage-specific marker, is used here as a negative control.
Figure 3
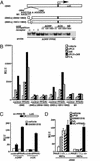
VLDL receptor is dramatically induced in PPARδ null macrophages. WT or _PPARδ_−/− ES-derived macrophages were treated with vehicle, rosiglitazone (1 μM), GW 501516 (0.1 μM), or VLDL (50 μg/ml) for 24 h. Total RNA (5 μg) was analyzed by Northern blotting using 32P-labeled cDNA probes. An equivalent amount of intact RNA was run present in each lane as accessed by hybridization to K-FABP cDNA probe.
Figure 4
VLDL and PPARδ regulate ADRP expression. (A) Lipid loading with VLDL/LPL results in redistribution of cytoplasmic ADRP. Expression vector containing HA-tagged mouse ADRP was transfected into CV-1 cells grown on chamber slides. After a 24-h treatment with VLDL/LPL, cells were stained with anti-HA antibody and visualized with FITC-conjugated secondary antibody. (B) PPARδ and VLDL regulate ADRP expression in macrophages. WT, PPARδ, or γ null macrophages were treated with cPGI (2 μM), GW 501516 (0.1 μM), or VLDL (25 μg/ml) for 24 h. (C) Ectopic expression of PPARδ confers VLDL responsiveness onto cells. NIH vector and NIH-PPARδ cells were treated with vehicle, cPGI (2 μM), LG 268 (0.1 μM), or VLDL (100 μg/ml) for 24 h. Gene expression analyses were performed by Northern blots with 32P-labeled cDNA probes.
Figure 5
ADRP promoter is a target for direct regulation by VLDL and PPARδ/RXR heterodimers. (A) ADRP promoter contains a PPRE. Schematic outline of reporter constructs containing the WT, point mutations, and internal deletions of the ADRP promoter. The potential PPRE is located between −2,004 and −1,992 bp of the ADRP promoter. Gel mobility shift assays were performed using in vitro translated proteins and 32P-end-labeled ADRP and acyl CoA oxidase (AoX) PPRE oligonucleotides. PPARα/RXR, PPARδ/RXR, and PPARγ/RXR heterodimers can bind to the ADRP PPRE. Unlabeled PPRE oligonucleotides or nonspecific DNA were used at the indicated molar excess to perform competition assays. Point mutations in the 5′ half site of the ADRP PPRE (M1) abolished binding of the PPARδ/RXR heterodimers. (B) PPARδ/RXR heterodimers transactivate the ADRP promoter. RAW 264.7 cells were cotransfected with ADRP reporter constructs (0.1 μg) and CMX-PPARδ/CMX-RXR expression vectors (10 ng) as described in Materials and Methods. Transfected cells were treated with ligands (2 μM cPGI, 0.1 μM LG 268, 2 μM cPGI + 0.1 μM LG 268, and VLDL (25 mg/ml)/LPL (10 mg/ml) for 24 h before collection for reporter gene assays. (C) The ADRP PPRE is a functional PPARδ response element. Three copies of the ADRP or the AOx PPRE were cloned upstream of the TK-luciferase reporter gene. CV-1 cells were cotransfected with reporters and receptor expression vectors and analyzed for luciferase activity as stated in B. (D) Endogenous PPARδ and VLDL transactivate the ADRP promoter. Transient transfection experiments were performed in _PPARα_−/− and _PPARδ_−/− MEFs with the luciferase reporter construct containing the proximal promoter (2,065 bp) of the ADRP gene.
Similar articles
- Activation of peroxisome proliferator-activated receptor δ inhibits human macrophage foam cell formation and the inflammatory response induced by very low-density lipoprotein.
Bojic LA, Sawyez CG, Telford DE, Edwards JY, Hegele RA, Huff MW. Bojic LA, et al. Arterioscler Thromb Vasc Biol. 2012 Dec;32(12):2919-28. doi: 10.1161/ATVBAHA.112.255208. Epub 2012 Sep 27. Arterioscler Thromb Vasc Biol. 2012. PMID: 23023367 - Effects of peroxisome proliferator-activated receptor delta on placentation, adiposity, and colorectal cancer.
Barak Y, Liao D, He W, Ong ES, Nelson MC, Olefsky JM, Boland R, Evans RM. Barak Y, et al. Proc Natl Acad Sci U S A. 2002 Jan 8;99(1):303-8. doi: 10.1073/pnas.012610299. Epub 2001 Dec 26. Proc Natl Acad Sci U S A. 2002. PMID: 11756685 Free PMC article. - Regulation of the growth arrest and DNA damage-inducible gene 45 (GADD45) by peroxisome proliferator-activated receptor gamma in vascular smooth muscle cells.
Bruemmer D, Yin F, Liu J, Berger JP, Sakai T, Blaschke F, Fleck E, Van Herle AJ, Forman BM, Law RE. Bruemmer D, et al. Circ Res. 2003 Aug 22;93(4):e38-47. doi: 10.1161/01.RES.0000088344.15288.E6. Epub 2003 Jul 24. Circ Res. 2003. PMID: 12881480 - [Peroxisome proliferator-activated receptor beta(delta) (PPAR beta(delta))].
Sugiyama E, Tanaka N, Aoyama T. Sugiyama E, et al. Nihon Rinsho. 2001 Feb;59 Suppl 2:301-4. Nihon Rinsho. 2001. PMID: 11351593 Review. Japanese. No abstract available.
Cited by
- Dose-dependent suppression of hyperlipidemia and intestinal polyp formation in Min mice by pioglitazone, a PPAR gamma ligand.
Niho N, Takahashi M, Shoji Y, Takeuchi Y, Matsubara S, Sugimura T, Wakabayashi K. Niho N, et al. Cancer Sci. 2003 Nov;94(11):960-4. doi: 10.1111/j.1349-7006.2003.tb01385.x. Cancer Sci. 2003. PMID: 14611672 Free PMC article. - Oxidative stress-induced FABP5 S-glutathionylation protects against acute lung injury by suppressing inflammation in macrophages.
Guo Y, Liu Y, Zhao S, Xu W, Li Y, Zhao P, Wang D, Cheng H, Ke Y, Zhang X. Guo Y, et al. Nat Commun. 2021 Dec 7;12(1):7094. doi: 10.1038/s41467-021-27428-9. Nat Commun. 2021. PMID: 34876574 Free PMC article. - PPAR-delta in Vascular Pathophysiology.
Wang N. Wang N. PPAR Res. 2008;2008:164163. doi: 10.1155/2008/164163. Epub 2009 Jan 6. PPAR Res. 2008. PMID: 19132133 Free PMC article. - PPARdelta regulates multiple proinflammatory pathways to suppress atherosclerosis.
Barish GD, Atkins AR, Downes M, Olson P, Chong LW, Nelson M, Zou Y, Hwang H, Kang H, Curtiss L, Evans RM, Lee CH. Barish GD, et al. Proc Natl Acad Sci U S A. 2008 Mar 18;105(11):4271-6. doi: 10.1073/pnas.0711875105. Epub 2008 Mar 12. Proc Natl Acad Sci U S A. 2008. PMID: 18337509 Free PMC article. - Bezafibrate at clinically relevant doses decreases serum/liver triglycerides via down-regulation of sterol regulatory element-binding protein-1c in mice: a novel peroxisome proliferator-activated receptor alpha-independent mechanism.
Nakajima T, Tanaka N, Kanbe H, Hara A, Kamijo Y, Zhang X, Gonzalez FJ, Aoyama T. Nakajima T, et al. Mol Pharmacol. 2009 Apr;75(4):782-92. doi: 10.1124/mol.108.052928. Epub 2009 Jan 5. Mol Pharmacol. 2009. PMID: 19124612 Free PMC article.
References
- Spiegelman B M, Flier J S. Cell. 2001;104:531–543. - PubMed
- Ross R. Am Heart J. 1999;138:419–420. - PubMed
- Glass C K, Witztum J L. Cell. 2001;104:503–516. - PubMed
- Nagy L, Tontonoz P, Alvarez J G, Chen H, Evans R M. Cell. 1998;93:229–240. - PubMed
- Janowski B A, Willy P J, Devi T R, Falck J R, Mangelsdorf D J. Nature. 1996;383:728–731. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases