Global expression profiling of fibroblast responses to transforming growth factor-beta1 reveals the induction of inhibitor of differentiation-1 and provides evidence of smooth muscle cell phenotypic switching - PubMed (original) (raw)
Global expression profiling of fibroblast responses to transforming growth factor-beta1 reveals the induction of inhibitor of differentiation-1 and provides evidence of smooth muscle cell phenotypic switching
Rachel C Chambers et al. Am J Pathol. 2003 Feb.
Abstract
Transforming growth factor-beta1 (TGF-beta1) plays a central role in promoting extracellular matrix protein deposition by promoting the transformation of fibroblasts to myofibroblasts. To gain new insights into the transcriptional programs involved, we profiled human fetal lung fibroblast global gene expression in response to TGF-beta1 up to 24 hours using oligonucleotide microarrays. In this report, we present data for 146 genes that were up-regulated at least twofold at two time points. These genes group into several major functional categories, including genes involved in cytoskeletal reorganization (n = 30), matrix formation (n = 25), metabolism and protein biosynthesis (n = 27), cell signaling (n = 21), proliferation and survival (n = 13), gene transcription (n = 9), and of uncertain function (n = 21). For 80 of these genes, this is the first report that they are TGF-beta1-responsive. The early induction of two members of the inhibitor of differentiation (ID) family of transcriptional regulators, ID1 and ID3, was followed by the up-regulation of a number of genes that are usually expressed by highly differentiated smooth muscle cells, including smooth muscle myosin heavy chain, basic calponin, and smoothelin. These findings were confirmed at the protein level for primary adult lung fibroblasts. ID1 further behaved like a typical immediate-early gene and, unlike ID3, was expressed and induced at the protein level. Immunohistochemical analysis showed that ID1 was highly expressed by (myo)fibroblasts within fibrotic foci in experimentally induced pulmonary fibrosis. ID1 acts as a dominant-negative antagonist of basic helix-loop-helix transcription factors that drive cell lineage commitment and differentiation. These findings have important implications for our understanding of fibroblast transcriptional programming in response to TGF-beta1 during development, oncogenesis, tissue repair, and fibrosis.
Figures
Figure 1.
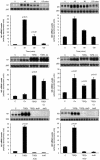
Global profile of fibroblast genes up-regulated in response to TGF-β1. Figure ▶ shows fold changes in gene expression over time for media control and TGF-β1-exposed cells. Genes are grouped into functional categories based on our current understanding of their most likely function as follows: A, genes encoding transcription factors; B, genes encoding signaling molecules; C, genes influencing cell survival and proliferation; D, genes associated with matrix formation; E, genes associated with cytoskeletal reorganization; F, genes involved in cell metabolism and protein synthesis. For each panel, each row displays expression data for a single gene, whereas columns represent pairwise comparisons in gene expression for TGF-β1-treatments relative to matching media controls at each time point as indicated by the labeling at the top of each panel; where TGF-β1.5, TGF-β6, TGF-β16, and TGF-β24 represent the pairwise comparison of the TGF-β1-treatments at 1-5, 6, 16, and 24 hours relative to the matching media control at each time point. Also shown are expression data for media controls, where C1.5, C6, C16, and C24 represents the comparison value for each time point relative to baseline expression (media control at 1.5 hours). Changes in gene expression are based on log-transformed values of the fold ratios of the signal intensities (referred to as average difference by Affymetrix) using a visual analog in which progressively brighter shades of red or green correspond to progressively greater gene inductions or repressions, respectively, as described by Eisen and colleagues. Genes are listed with their GenBank accession codes, followed by their symbols and full gene name according to the HUGO nomenclature. An asterisk denotes that the gene is a newly identified TGF-β-responsive gene. Genes were selected on the basis of at least a twofold increase in gene expression at two time points in response to TGF-β1 treatment compared with the corresponding media controls. Also included are three genes encoding signaling molecules (ARHB, MAP2K1, RAC1) and four genes encoding transcription factors (C-MYC, FOSL2, HRY, ID3), which were increased by at least fivefold at a single time point.
Figure 2.
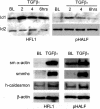
ID1 and ID3 behave like direct TGF-β1 target genes. Top: The effect of TGF-β1 on ID1 and ID3 mRNA levels over time assessed by Northern analysis. Middle: The effect of cycloheximide on TGF-β1-induced ID1 and ID3 mRNA levels. Bottom: The effect of TGF-β1 on ID1 and ID3 mRNA levels in cells pretreated with actinomycin D. Data for HFL-1 fibroblasts fetal is shown on the left; whereas the right shows data for primary adult lung fibroblasts (pHALF). Bar graphs represent the mean of three replicate cultures. Also shown are representative phosphorimages of the ID1 and ID3 transcripts and corresponding images of the 28S rRNA bands. P values represent comparisons made to serum-free media control-treated cells.
Figure 3.
TGF-β induces the expression of ID1 and smooth muscle cell differentiation marker proteins. Top: A representative Western blot for the effect of TGF-β1 in serum-free conditions on ID1 protein levels up to 6 hours for human fetal lung fibroblasts (HFL1) and primary human adult lung fibroblasts (pHALF) compared with baseline (BL). Also shown are ID2 protein levels that did not change on exposure to TGF-β1. In contrast, ID3 was undetectable at the protein level. Bottom: Representative Western blots for the effect of TGF-β1 for 36 hours on the indicated proteins for cultures of HFL-1 and pHALF compared with cultures exposed to serum-free control media (BL).
Figure 4.
Effect of TGF-β1 on smooth muscle differentiation marker protein expression in fetal and primary adult lung fibroblasts by immunocytofluorescence. Images show human fetal lung fibroblasts (HFL-1) (first two columns) and primary human adult lung fibroblasts (pHALF) (last two columns) exposed to serum-free control media or TGF-β1 for 36 hours and stained for the indicated proteins by immunocytofluorescence. Images are representative of a minimum of 20 fields viewed at high magnification (×1000 under oil immersion) for three separate experiments performed. a–d: Smooth muscle α-actin. a, HFL1 in control media; b, HFL1 and TGF-β1; c, pHALF in control media; d, pHALF and TGF-β1. e–h: Smooth muscle myosin heavy chain. e, HFL1 in control media; f, HFL1 and TGF-β1; g, pHALF in control media; h, pHALF and TGF-β1. i–l: h-caldesmon. i, HFL1 in control media; j, HFL1 and TGF-β1; k, pHALF in control media; l, pHALF and TGF-β1.m–p: Smoothelin. m, HFL1 in control media; n, HFL1 and TGF-β1; o, pHALF in control media; p, pHALF and TGF-β1.
Figure 5.
Immunohistochemical analysis of ID1 expression in experimental pulmonary fibrosis. Figure ▶ shows brown immunoperoxidase staining for ID1 in insufflated normal rat lung tissue 14 days after intratracheal instillation of saline (SAL) compared with rats given bleomycin (Bleo). In the normal lung, ID1 immunoreactivity is confined to smooth muscle bundles surrounding major airways (A) and blood vessels (V) but in bleomycin-induced pulmonary fibrosis, ID1 was also immunolocalized to (myo)fibroblasts within fibrotic foci, with some cells showing nuclear staining (arrow). Original magnifications, ×400; counterstained with Mayers hematoxylin.
Similar articles
- Role of inhibitor of differentiation 3 gene in cellular differentiation of human corneal stromal fibroblasts.
Gupta S, Martin LM, Sinha NR, Smith KE, Sinha PR, Dailey EM, Hesemann NP, Mohan RR. Gupta S, et al. Mol Vis. 2020 Nov 25;26:742-756. eCollection 2020. Mol Vis. 2020. PMID: 33273801 Free PMC article. - Hormonal regulation and differential actions of the helix-loop-helix transcriptional inhibitors of differentiation (Id1, Id2, Id3, and Id4) in Sertoli cells.
Chaudhary J, Johnson J, Kim G, Skinner MK. Chaudhary J, et al. Endocrinology. 2001 May;142(5):1727-36. doi: 10.1210/endo.142.5.8134. Endocrinology. 2001. PMID: 11316735 - TGF-beta 1 induces proliferation in human renal fibroblasts via induction of basic fibroblast growth factor (FGF-2).
Strutz F, Zeisberg M, Renziehausen A, Raschke B, Becker V, van Kooten C, Müller G. Strutz F, et al. Kidney Int. 2001 Feb;59(2):579-92. doi: 10.1046/j.1523-1755.2001.059002579.x. Kidney Int. 2001. PMID: 11168939 - Transcriptional regulation of metastatic [Id]entity by KLF17.
Iwanicki MP, Brugge JS. Iwanicki MP, et al. Genome Biol. 2009;10(11):244. doi: 10.1186/gb-2009-10-11-244. Epub 2009 Nov 30. Genome Biol. 2009. PMID: 19951400 Free PMC article. Review. - Novel fibroblast phenotypes in homeostasis and chronic inflammation: From functions to potential regulators.
Miki H, Manresa MC. Miki H, et al. J Physiol. 2023 Jun;601(12):2273-2291. doi: 10.1113/JP284620. Epub 2023 May 2. J Physiol. 2023. PMID: 37062932 Review.
Cited by
- Cardiac fibrosis: Myofibroblast-mediated pathological regulation and drug delivery strategies.
Liu M, López de Juan Abad B, Cheng K. Liu M, et al. Adv Drug Deliv Rev. 2021 Jun;173:504-519. doi: 10.1016/j.addr.2021.03.021. Epub 2021 Apr 5. Adv Drug Deliv Rev. 2021. PMID: 33831476 Free PMC article. Review. - Alterations in the Smad pathway in human cancers.
Samanta D, Datta PK. Samanta D, et al. Front Biosci (Landmark Ed). 2012 Jan 1;17(4):1281-93. doi: 10.2741/3986. Front Biosci (Landmark Ed). 2012. PMID: 22201803 Free PMC article. Review. - Genomic deregulation during metastasis of renal cell carcinoma implements a myofibroblast-like program of gene expression.
López-Lago MA, Thodima VJ, Guttapalli A, Chan T, Heguy A, Molina AM, Reuter VE, Motzer RJ, Chaganti RS. López-Lago MA, et al. Cancer Res. 2010 Dec 1;70(23):9682-92. doi: 10.1158/0008-5472.CAN-10-2279. Epub 2010 Oct 15. Cancer Res. 2010. PMID: 20952505 Free PMC article. - Aging Effects on Optic Nerve Neurodegeneration.
Coleman-Belin J, Harris A, Chen B, Zhou J, Ciulla T, Verticchio A, Antman G, Chang M, Siesky B. Coleman-Belin J, et al. Int J Mol Sci. 2023 Jan 29;24(3):2573. doi: 10.3390/ijms24032573. Int J Mol Sci. 2023. PMID: 36768896 Free PMC article. Review. - Dosage Effects of an 810 nm Diode Laser on the Proliferation and Growth Factor Expression of Human Gingival Fibroblasts.
Karoussis IK, Kyriakidou K, Psarros C, Afouxenides P, Vrotsos IA. Karoussis IK, et al. J Lasers Med Sci. 2021 Jun 20;12:e25. doi: 10.34172/jlms.2021.25. eCollection 2021. J Lasers Med Sci. 2021. PMID: 34733748 Free PMC article.
References
- Massague J, Blain SW, Lo RS: TGFbeta signaling in growth control, cancer, and heritable disorders. Cell 2000, 103:295-309 - PubMed
- Blobe GC, Schiemann WP, Lodish HF: Role of transforming growth factor beta in human disease. N Engl J Med 2000, 342:1350-1358 - PubMed
- Wrana JL, Attisano L, Wieser R, Ventura F, Massague J: Mechanism of activation of the TGF-beta receptor. Nature 1994, 370:341-347 - PubMed
- Heldin CH, Miyazono K, ten Dijke P: TGF-beta signalling from cell membrane to nucleus through SMAD proteins. Nature 1997, 390:465-471 - PubMed
- Massague J: TGF-beta signal transduction. Annu Rev Biochem 1998, 67:753-791 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous