Drosophila p120catenin plays a supporting role in cell adhesion but is not an essential adherens junction component - PubMed (original) (raw)
Drosophila p120catenin plays a supporting role in cell adhesion but is not an essential adherens junction component
Steven H Myster et al. J Cell Biol. 2003.
Abstract
Cadherin-catenin complexes, localized to adherens junctions, are essential for cell-cell adhesion. One means of regulating adhesion is through the juxtamembrane domain of the cadherin cytoplasmic tail. This region is the binding site for p120, leading to the hypothesis that p120 is a key regulator of cell adhesion. p120 has also been suggested to regulate the GTPase Rho and to regulate transcription via its binding partner Kaiso. To test these hypothesized functions, we turned to Drosophila, which has only a single p120 family member. It localizes to adherens junctions and binds the juxtamembrane region of DE-cadherin (DE-cad). We generated null alleles of p120 and found that mutants are viable and fertile and have no substantial changes in junction structure or function. However, p120 mutations strongly enhance mutations in the genes encoding DE-cadherin or Armadillo, the beta-catenin homologue. Finally, we examined the localization of p120 during embryogenesis. p120 localizes to adherens junctions, but its localization there is less universal than that of core adherens junction proteins. Together, these data suggest that p120 is an important positive modulator of adhesion but that it is not an essential core component of adherens junctions.
Figures
Figure 1.
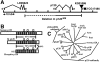
Drosophila p120 is a member of the p120 subfamily. (A) Gene structure of p120 and the two adjacent genes, LD05623 and CG17486. KG01086, the P element insertion used to generate p120 mutants, is indicated, as is the region deleted in p120 308 (uncertainty in the left boundary is indicated as a dotted line). (B) Human, fly, and mosquito p120. Gray boxes represent Arm repeats. Repeat 6, which diverges from the consensus, is indicated by a “?”. Loops represent conserved inserts in Arm repeats. Hatched box shows conserved region of similarity. Amino acid identities in pairwise comparisons of the regions bracketed are indicated. (C) Unrooted tree of the p120 subfamily, plakophilin subfamily, and selected other Arm repeat proteins. H, human; M, mouse; X, Xenopus.
Figure 2.
p120 localizes to AJs and the cytoplasm. (A and B) Wild-type (WT) and homozygous p120 308 (p120) embryonic extracts immunoblotted with affinity-purified rat anti-p120 (A) or rabbit anti-p120 (B). (C–O) Embryos. Embryonic stages are as in Wieschaus and Nüsslein-Volhard (1986). (C and D) Stage 11. (C) Apical section through epidermis; more basal section, cutting across the folded epithelium (D). p120 localizes to the apical cell cortex (arrows). (E–G) Stage 11. p120 (red); Arm (green). Cells are indicated in which p120 accumulation is relatively high (red arrows) or low (green arrow). (H) Stage 11 p120 380 mutant. (I) Live image; p120-GFP. Higher levels are seen at the ends of cells that are stretched (arrows). (J) Stage 12, ubiquitin-myc-p120. (K and L) Stage 12, expressing myc-p120 in prd stripes. Myc (red), DE-cad (green). DE-cad is uniform across the embryo; thus, myc-p120 overexpression does not affect its localization. (M–O) Stage 17, optical cross sections. (M and N) p120 (red) and Arm (green) colocalize to apical AJs of the gut (white arrows). (O) p120-GFP (green) and DE-cad (red) colocalize at AJs of the gut (arrow) and epidermis (arrowhead). (P) Egg chamber in ovary. p120-GFP (green) and Arm (red) colocalize at follicle cell AJs (arrow). Bars, 5 μm.
Figure 3.
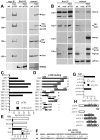
p120 is in AJ complexes and binds the JM region of DE-cad. (A) Cell extracts or IPs using anti-myc or anti-BicD (negative control) from wild-type or myc-p120 embryos were immunoblotted with antibodies against myc, p120, DE-cad, Arm, and BicD. Proteins identified are indicated to the right, and selected mol wt markers (kD) are on the left. Rat anti-p120 recognizes both endogenous and myc-p120. (B) Cell extracts or anti-Arm IPs from wild-type (wt), p120 mutant (mut), and myc-p120 expressing embryos were immunoblotted with antibodies against Arm, DE-cad, myc, p120, and Pnut (a negative control). (A and B) ∼1% of extract and ∼50% of each IP was loaded. (C–H) Yeast two-hybrid interactions assessed by β-galactosidase activity. (C) Interaction between p120 and the DE-cad cytoplasmic tail (DEC), DE-cad deletion constructs (DECXX; black bars) or vector control (white bars). (D) Schematic illustrating DEC deletion constructs in C. (E and F) Clustered point mutations in the JM region of the DE-cad cytoplasmic tail (DECM5 and DECM6, diagram in F; dE, DE-cad; mE, mouse E-cad) abolished binding of full-length p120 (E, black bar), but had no effect on interaction of the Arm repeats of Arm with DE-cad (E, white bars). (G and H) Arm repeats 1–10 of p120 are required to confer strong binding to DE-cad (black bars). Vector control (white bars). (H) Diagram of constructs used.
Figure 4.
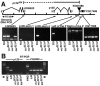
_p120_308 is a null allele. (A) p120 308 deletes the entire p120 coding region, but does not affect other genes. Schematic as in Fig. 1 A. Genomic DNA from single wild-type or homozygous mutant flies was PCR amplified using primer pairs from the indicated regions between LD05623 and CG17486. (B) The p120 mutants are mRNA nulls. cDNA generated from oligo-dT-primed total RNA from p120 mutants and wild-type was amplified with primers spanning the p120 third intron. An unrelated gene, CG2905, is a control. A DNA control confirmed we were examining mRNA.
Figure 5.
AJ proteins are not significantly altered in levels or localization in p120 mutants. (A) Embryonic extracts from 3–8 h wild-type (WT) and p120 308 (mut) strains immunoblotted with antibodies to p120 (arrowhead), DE-cad, α-cat, and Arm. Anti-Pnut is a loading control. Mol wt standards (kD) are at left. (B–O) Wild-type and p120 308 mutant embryos labeled with Arm (B–G), DE-cad (H and I), α-cat (J and K), or phalloidin to show F-actin (L–O). (F, G, N, and O) arrows indicate the leading edge during dorsal closure. Bars, 5 μm.
Figure 6.
p120 mutations strongly enhance shg and arm. Cuticle preps, anterior up. (A) Wild-type. Note alternating denticle bands and naked cuticle on the ventral epidermis and normal head exoskeleton (top). (B) shg g119. Note head involution defects (arrow) but intact ventral epidermis. (C) Zygotic shg g119 p120 308. Note hole in ventral epidermis. (D) Zygotic shg g119 p120 308 mutant that is also maternally p120 308 mutant. The entire ventral epidermis is lost. (E) shg R69. Ventral epidermis is lost, but dorsal and lateral epidermis remain. (F) Zygotic shg R69 p120 308. Dorsal epidermis is disrupted (arrow). (G) Zygotic shg R69 p120 308 mutant that was also maternally p120 308 mutant. The remaining cuticle is fragmented. (H) shg 2. Note holes in ventral epidermis. (I) Zygotic shg 2 p120 308. Note complete loss of ventral epidermis. (J) arm YD35. Note shortened body and lawn of denticles ventrally (arrowhead), and defects in dorsal closure (arrow). (K and L) Zygotic arm YD35 p120 308 mutant that was also maternally p120 308 mutant. Cuticles are longer, and in L dorsal closure defects are suppressed.
Figure 7.
p120 mRNA is ubiquitously expressed but enriched in certain tissues. Embryos at indicated stages, probed by situ hybridization for expression of p120 mRNA (A–D and F) or with a sense strand p120 control probe (E). Anterior is to the left and dorsal is up. In B, white arrows indicate the neurectoderm and mesoderm of the germband. Certain tissues accumulate elevated levels of p120 mRNA, e.g., the posterior midgut (B, black arrow), brain and CNS (C and F, white arrows), migrating anterior and posterior midgut (C, black arrows), cells forming the midgut constriction (D, arrow). (F) Stage 14 wild-type and mutant homozygous for a deletion removing p120 (Df[2R]M41A8), showing remaining maternal mRNA.
Figure 8.
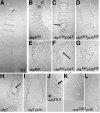
p120 localization during the syncytial development and cellularization differs from that of Arm. (A–J) Syncytial blastoderm (A and B) and cellularizing embryos (C–J). p120 (red); Arm (green). (A–D, G, and H) Surface sections. (E, F, I, and J) Optical cross sections. (A and B) Arm localizes strongly to pseudocleavage furrows, whereas p120 staining is much less intense (arrow). (C–F) Early cellularization. p120 colocalizes with Arm at basal junctions (E and F, arrows) and also stains paired structures in the cytoplasm (C and D, arrows). (G–J) Mid-late cellularization. p120 is reduced at cell junctions (H, arrow) compared with Arm (G). Arm labels basal junctions, lateral membranes, and nascent AJs (I). p120 localizes to an apical domain (J, arrow). (K–M and O–S) Embryos expressing p120-GFP. (K–M) Live images, cellularization. p120-GFP localizes to basal junctions (K and M, arrowhead), nascent AJs (L), and centrosomes (L and M arrows). (N) Syncytial embryo, rabbit anti-p120. (O and P) p120-GFP (green), propidium iodide to label DNA (red), γ-tubulin (gtub, blue). p120-GFP and γ-tubulin colocalize (arrow) and p120-GFP is on mitotic spindles (arrowhead). (Q–S) p120-GFP (green); centrosomin (cnn, red). p120-GFP is enriched at cell junctions (arrowhead) and with condensing DNA (black arrow), and colocalizes with centrosomin (white arrow). Centrosomal p120-GFP is absent by stage 9 (S). Bars, 5 μm.
Figure 9.
p120 localizes to embryonic cell junctions but is not as uniformly distributed as core AJ proteins. (A–D) Stage 8. p120 (red); Arm (green). (Arrows) Cell junctions accumulating both p120 and Arm; (arrowheads) cell junctions depleted for p120. (E) Stage 9. p120 junctional accumulation is becoming more uniform. In mitotic cells, it is absent from the cytoplasm (arrows). (F–H) Stage 9. Histone-GFP (green); p120 (red); microtubules (MT; blue). (F and G) Apical planes; (H) section through the middle of the cells. In nonmitotic cells, p120 localizes in the cytoplasm and at cell junctions (F and G, red arrow). In mitotic cells, junctional and cytoplasmic p120 is reduced (E, arrows; F–H white arrows). Some junctional staining remains at the cell mid-plane (H, red arrow). (I) Stage 12/13. (J–N) Stage 14. p120 (red); Arm (green). (O) Stage 14. Live image, p120-GFP. (Arrows) Accumulation at the ends of stretched cells. Bars, 5 μm.
Figure 10.
p120 colocalizes with Arm in other tissues. (A–D) Stage 15. p120 (red); Arm (green). Arm is enriched in fusion cells (red arrows). (E) p120-GFP. (F and G) Stage 15–16. p120 (red) and Arm (green) colocalize to peripheral nervous system chordotonal organs. (F and G, insets) Similar stage p120 mutant. Arm localizes to chordotonals; p120 is lost. (H) p120-GFP in chordotonals. (I–K) Stage 17. p120 (red); Arm (green). (L) p120-GFP. (M–N) p120-GFP. (M) Eye imaginal disc. Undifferentiated cells (arrowheads), photoreceptor cells (arrows). (N and O) Larval brain. (N) p120-GFP at cell borders between neuroblasts (arrowheads) and ganglion mother cells (arrow). (O) Progeny of neuroblasts (arrowheads) sending bundled axons (arrows) to the neuropil. (P) Stage 10 egg chamber. p120-GFP (green) and Arm (red) colocalize at cell junctions of migrating border cells (arrow). In nurse cells, p120-GFP is cytoplasmic but excluded from nuclei (arrowhead). Bars, 5 μm.
Similar articles
- Binding site for p120/delta-catenin is not required for Drosophila E-cadherin function in vivo.
Pacquelet A, Lin L, Rorth P. Pacquelet A, et al. J Cell Biol. 2003 Feb 3;160(3):313-9. doi: 10.1083/jcb.200207160. Epub 2003 Jan 27. J Cell Biol. 2003. PMID: 12551956 Free PMC article. - The Caenorhabditis elegans p120 catenin homologue, JAC-1, modulates cadherin-catenin function during epidermal morphogenesis.
Pettitt J, Cox EA, Broadbent ID, Flett A, Hardin J. Pettitt J, et al. J Cell Biol. 2003 Jul 7;162(1):15-22. doi: 10.1083/jcb.200212136. J Cell Biol. 2003. PMID: 12847081 Free PMC article. - A core function for p120-catenin in cadherin turnover.
Davis MA, Ireton RC, Reynolds AB. Davis MA, et al. J Cell Biol. 2003 Nov 10;163(3):525-34. doi: 10.1083/jcb.200307111. J Cell Biol. 2003. PMID: 14610055 Free PMC article. - Delta-catenin at the synaptic-adherens junction.
Kosik KS, Donahue CP, Israely I, Liu X, Ochiishi T. Kosik KS, et al. Trends Cell Biol. 2005 Mar;15(3):172-8. doi: 10.1016/j.tcb.2005.01.004. Trends Cell Biol. 2005. PMID: 15752981 Review. - Adherens junction assembly and function in the Drosophila embryo.
Harris TJ. Harris TJ. Int Rev Cell Mol Biol. 2012;293:45-83. doi: 10.1016/B978-0-12-394304-0.00007-5. Int Rev Cell Mol Biol. 2012. PMID: 22251558 Review.
Cited by
- RhoGAP19D inhibits Cdc42 laterally to control epithelial cell shape and prevent invasion.
Fic W, Bastock R, Raimondi F, Los E, Inoue Y, Gallop JL, Russell RB, St Johnston D. Fic W, et al. J Cell Biol. 2021 Apr 5;220(4):e202009116. doi: 10.1083/jcb.202009116. J Cell Biol. 2021. PMID: 33646271 Free PMC article. - Adherens junction turnover: regulating adhesion through cadherin endocytosis, degradation, and recycling.
Kowalczyk AP, Nanes BA. Kowalczyk AP, et al. Subcell Biochem. 2012;60:197-222. doi: 10.1007/978-94-007-4186-7_9. Subcell Biochem. 2012. PMID: 22674073 Free PMC article. Review. - Dynamic contacts: rearranging adherens junctions to drive epithelial remodelling.
Takeichi M. Takeichi M. Nat Rev Mol Cell Biol. 2014 Jun;15(6):397-410. doi: 10.1038/nrm3802. Epub 2014 May 14. Nat Rev Mol Cell Biol. 2014. PMID: 24824068 Review. - Drosophila PATJ supports adherens junction stability by modulating Myosin light chain activity.
Sen A, Nagy-Zsvér-Vadas Z, Krahn MP. Sen A, et al. J Cell Biol. 2012 Nov 12;199(4):685-98. doi: 10.1083/jcb.201206064. Epub 2012 Nov 5. J Cell Biol. 2012. PMID: 23128243 Free PMC article. - p120 catenin is required for the stress response in Drosophila.
Stefanatos RK, Bauer C, Vidal M. Stefanatos RK, et al. PLoS One. 2013 Dec 12;8(12):e83942. doi: 10.1371/journal.pone.0083942. eCollection 2013. PLoS One. 2013. PMID: 24349561 Free PMC article.
References
- Akong, K., B. McCartney, and M. Peifer. 2002. Drosophila APC2 and APC1 have overlapping roles in the larval brain despite their distinct intracellular localizations. Dev. Biol. 250:71–90. - PubMed
- Anastasiadis, P.Z., and P.B. Reynolds. 2000. The p120 catenin family: complex roles in adhesion, signaling and cancer. J. Cell Sci. 113:1319–1334. - PubMed
- Anastasiadis, P.Z., S.Y. Moon, M.A. Thoreson, D.J. Mariner, H.C. Crawford, Y. Zheng, and A.B. Reynolds. 2000. Inhibition of RhoA by p120 catenin. Nat. Cell Biol. 2:637–644. - PubMed
- Baki, L., P. Marambaud, S. Efthimiopoulos, A. Georgakopoulos, P. Wen, W. Cui, J. Shioi, E. Koo, M. Ozawa, V.L. Friedrich, Jr., and N.K. Robakis. 2001. Presenilin-1 binds cytoplasmic epithelial cadherin, inhibits cadherin/p120 association, and regulates stability and function of the cadherin/catenin adhesion complex. Proc. Natl. Acad. Sci. USA. 98:2381–2386. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous