Determination of the transcriptome of Vibrio cholerae during intraintestinal growth and midexponential phase in vitro - PubMed (original) (raw)
Determination of the transcriptome of Vibrio cholerae during intraintestinal growth and midexponential phase in vitro
Qing Xu et al. Proc Natl Acad Sci U S A. 2003.
Abstract
Vibrio cholerae is the etiologic bacterial agent of cholera, a severe diarrheal disease endemic in much of the developing world. The V. cholerae genome contains 3,890 genes distributed between a large and a small chromosome. Although the large chromosome encodes the majority of recognizable gene products and virulence determinants, the small chromosome carries a disproportionate number of hypothetical genes. Thus, little is known about the role of the small chromosome in the biology of this organism or other Vibrio species. We have used the rabbit ileal loop model of V. cholerae infection to obtain in vivo-grown cells under near midexponential conditions in the small-intestinal environment. We compared the global transcriptional pattern of these in vivo-grown cells to those grown to midexponential phase in rich medium under aerobic conditions. Under both conditions, the genes showing the highest levels of expression reside primarily on the large chromosome. However, a shift occurs in vivo that results in many more small chromosomal genes being expressed during growth in the intestine. Our analysis further suggests that nutrient limitation (particularly iron) and anaerobiosis are major stresses experienced by V. cholerae during growth in the rabbit upper intestine. Finally, relative to in vitro growth, the intestinal environment significantly enhanced expression of several virulence genes, including those involved in phenotypes such as motility, chemotaxis, intestinal colonization, and toxin production.
Figures
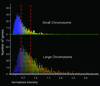
Figure 1
Graphical representation of V. cholerae gene expression in LB. (Upper) Distribution of small-chromosome genes. (Lower) Distribution of large-chromosome genes. Three thousand eight hundred-ninety genes were analyzed by using GENESPRING, and the expression levels of these genes are represented by normalized intensities. On the basis of expression levels, dashed lines divide the total genes into three equal areas: top one-third, middle one-third, and bottom one-third. One hundred seven genes, whose expression levels were >3.5, are not listed.
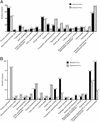
Figure 2
Functional classes of differentially expressed V. cholerae genes. (A) Functional categories of the 300 genes with the highest expression levels in vitro (aerobic growth in LB) and in vivo (growth in rabbit ileal loops). (B) Functional categories of genes showing 2-fold or greater changes under in vivo compared with in vitro growth conditions. The percentage of genes in each category appears above each bar.
Figure 3
RT-PCR of total RNA isolated from rabbit ileal loops. PCR was performed as described in Materials and Methods by using primers specific for selected V. cholerae genes and either reverse-transcribed RNA (cDNA) or chromosomal DNA as template.
Similar articles
- Aerobic Metabolism in Vibrio cholerae Is Required for Population Expansion during Infection.
Van Alst AJ, DiRita VJ. Van Alst AJ, et al. mBio. 2020 Sep 1;11(5):e01989-20. doi: 10.1128/mBio.01989-20. mBio. 2020. PMID: 32873763 Free PMC article. - RpoS controls the Vibrio cholerae mucosal escape response.
Nielsen AT, Dolganov NA, Otto G, Miller MC, Wu CY, Schoolnik GK. Nielsen AT, et al. PLoS Pathog. 2006 Oct;2(10):e109. doi: 10.1371/journal.ppat.0020109. PLoS Pathog. 2006. PMID: 17054394 Free PMC article. - Mutation in the relA gene of Vibrio cholerae affects in vitro and in vivo expression of virulence factors.
Haralalka S, Nandi S, Bhadra RK. Haralalka S, et al. J Bacteriol. 2003 Aug;185(16):4672-82. doi: 10.1128/JB.185.16.4672-4682.2003. J Bacteriol. 2003. PMID: 12896985 Free PMC article. - Multiple intraintestinal signals coordinate the regulation of Vibrio cholerae virulence determinants.
Peterson KM, Gellings PS. Peterson KM, et al. Pathog Dis. 2018 Feb 1;76(1). doi: 10.1093/femspd/ftx126. Pathog Dis. 2018. PMID: 29315383 Review. - Expression of Vibrio cholerae virulence genes in response to environmental signals.
Peterson KM. Peterson KM. Curr Issues Intest Microbiol. 2002 Sep;3(2):29-38. Curr Issues Intest Microbiol. 2002. PMID: 12400636 Review.
Cited by
- Promotion of colonization and virulence by cholera toxin is dependent on neutrophils.
Queen J, Satchell KJ. Queen J, et al. Infect Immun. 2013 Sep;81(9):3338-45. doi: 10.1128/IAI.00422-13. Epub 2013 Jun 24. Infect Immun. 2013. PMID: 23798539 Free PMC article. - The two chromosomes of Vibrio cholerae are initiated at different time points in the cell cycle.
Rasmussen T, Jensen RB, Skovgaard O. Rasmussen T, et al. EMBO J. 2007 Jul 11;26(13):3124-31. doi: 10.1038/sj.emboj.7601747. Epub 2007 Jun 7. EMBO J. 2007. PMID: 17557077 Free PMC article. - A novel role for enzyme I of the Vibrio cholerae phosphoenolpyruvate phosphotransferase system in regulation of growth in a biofilm.
Houot L, Watnick PI. Houot L, et al. J Bacteriol. 2008 Jan;190(1):311-20. doi: 10.1128/JB.01410-07. Epub 2007 Nov 2. J Bacteriol. 2008. PMID: 17981973 Free PMC article. - Transcriptome changes in Mycoplasma hyopneumoniae during infection.
Madsen ML, Puttamreddy S, Thacker EL, Carruthers MD, Minion FC. Madsen ML, et al. Infect Immun. 2008 Feb;76(2):658-63. doi: 10.1128/IAI.01291-07. Epub 2007 Dec 10. Infect Immun. 2008. PMID: 18070898 Free PMC article. - Genetics-squared: combining host and pathogen genetics in the analysis of innate immunity and bacterial virulence.
Persson J, Vance RE. Persson J, et al. Immunogenetics. 2007 Oct;59(10):761-78. doi: 10.1007/s00251-007-0248-0. Epub 2007 Sep 14. Immunogenetics. 2007. PMID: 17874090 Review.
References
- Waldor M K, Mekalanos J J. Science. 1996;272:1910–1914. - PubMed
- Cotter P A, DiRita V J. Annu Rev Microbiol. 2000;54:519–565. - PubMed
- Lee S H, Hava D L, Waldor M K, Camilli A. Cell. 1999;99:625–634. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases