A novel algorithm for computational identification of contaminated EST libraries - PubMed (original) (raw)
A novel algorithm for computational identification of contaminated EST libraries
Rotem Sorek et al. Nucleic Acids Res. 2003.
Abstract
A key goal of the Human Genome Project was to understand the complete set of human proteins, the proteome. Since the genome sequence by itself is not sufficient for predicting new genes and alternative splicing events that lead to new proteins, expressed sequence tags (ESTs) are used as the primary tool for these purposes. The high prevalence of artifacts in dbEST, however, often leads to invalid predictions. Here we describe a novel method for recognizing genomic DNA contamination and other artifacts that cannot be identified using current EST cleaning techniques. Our method uses the alignment of the entire set of ESTs to the human genome to identify highly contaminated EST libraries. We discovered 53 highly contaminated libraries and a subset of 24 766 ESTs from these libraries that probably represent contamination with genomic DNA, pre-mRNA, and ESTs that span non-canonical introns. Although this is only a small fraction of the entire EST dataset, each contaminating sequence could create a spurious transcript prediction. Indeed, in the clustering and assembly tool that we used, these sequences would have caused incorrect inference of 9575 new splice variants and 6370 new genes. Conclusions based on EST analysis, including prediction of alternative splicing, should be re-evaluated in light of these results. Our method, along with the identified set of contaminated sequences, will be essential for applications that depend on large EST datasets.
Figures
Figure 1
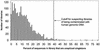
Number of EST libraries as a function of the percentage of unspliced singletons in the library. The percentage of unspliced singletons in each of the 1906 libraries was computed using the LEADS clustering and assembly tool. The mean percentage was 11.3%, with a standard deviation of 9.3%. The cut-off indicated by the arrow is three standard deviations above the mean, or 39.3%. The 21 libraries with percentages above this limit are probably highly contaminated with human genomic DNA.
Figure 2
A cluster that contains a sequence suspected of being human genomic DNA contamination. Although most ESTs that represent genomic DNA come from intergenic regions, some appear in clusters that contain other sequences. This figure shows an example of such a case. The two dark blue lines at the top represent predicted transcripts, the red line below them represents the genome, and the three following black and purple lines represent ESTs. The second EST (black bar) represents sequence AA601679 from EST library NCI_CGAP_PHE1, which was identified as having a high rate of genomic DNA contamination. The extension of the black bar to the left of the purple bar above it (denoted by the thin black arrow) is probably a portion of an intron that was included because of genomic DNA contamination. This resulted in the prediction of an additional transcript (transcript_0) that is likely to be spurious.
Figure 3
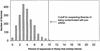
Number of EST libraries as a function of the percentage of sequences in the library that overlap introns. The percentage of sequences that overlap introns in each of the 1906 libraries was computed using the LEADS clustering and assembly tool. The mean percentage was 3.7%, with a standard deviation of 2.0%. The cut-off indicated by the arrow is three standard deviations above the mean, or 9.8%. The 14 libraries with percentages above this limit are probably highly contaminated with pre-mRNA sequences.
Figure 4
A cluster that exhibits a non-canonical intron. The two dark blue lines at the top represent predicted transcripts, the red line below them represents the genome, and the six following purple lines represent ESTs. The gap indicated by the black arrow begins with AA and ends with TT (i.e., it is a non-canonical intron). The gap is 54 bases long, which is similar to many of the observed non-canonical introns found in problematic libraries. The gap occurred only in the sequence that came from a library detected as possibly contaminated; it may correspond to sequence that was incorrectly deleted from some ESTs. This gap resulted in the prediction of a splice variant (transcript_1) that is likely spurious.
Similar articles
- Gene structure prediction and alternative splicing analysis using genomically aligned ESTs.
Kan Z, Rouchka EC, Gish WR, States DJ. Kan Z, et al. Genome Res. 2001 May;11(5):889-900. doi: 10.1101/gr.155001. Genome Res. 2001. PMID: 11337482 Free PMC article. - A comparison of expressed sequence tags (ESTs) to human genomic sequences.
Wolfsberg TG, Landsman D. Wolfsberg TG, et al. Nucleic Acids Res. 1997 Apr 15;25(8):1626-32. doi: 10.1093/nar/25.8.1626. Nucleic Acids Res. 1997. PMID: 9092672 Free PMC article. - How prevalent is functional alternative splicing in the human genome?
Sorek R, Shamir R, Ast G. Sorek R, et al. Trends Genet. 2004 Feb;20(2):68-71. doi: 10.1016/j.tig.2003.12.004. Trends Genet. 2004. PMID: 14746986 Review. - Reconstruction of full-length isoforms from splice graphs.
Xing Y, Lee C. Xing Y, et al. Methods Mol Biol. 2008;452:199-205. doi: 10.1007/978-1-60327-159-2_10. Methods Mol Biol. 2008. PMID: 18566766 Review.
Cited by
- A non-EST-based method for exon-skipping prediction.
Sorek R, Shemesh R, Cohen Y, Basechess O, Ast G, Shamir R. Sorek R, et al. Genome Res. 2004 Aug;14(8):1617-23. doi: 10.1101/gr.2572604. Genome Res. 2004. PMID: 15289480 Free PMC article. - Under-representation of PolyA/PolyT tailed ESTs in human ESTdb: an obstacle to alternative polyadenylation inference.
Gilat R, Goncharov S, Esterman N, Shweiki D. Gilat R, et al. Bioinformation. 2006 Oct 7;1(6):220-4. Bioinformation. 2006. PMID: 17597892 Free PMC article. - Computational comparative analyses of alternative splicing regulation using full-length cDNA of various eukaryotes.
Itoh H, Washio T, Tomita M. Itoh H, et al. RNA. 2004 Jul;10(7):1005-18. doi: 10.1261/rna.5221604. RNA. 2004. PMID: 15208437 Free PMC article. - Simultaneous identification of long similar substrings in large sets of sequences.
Kleffe J, Möller F, Wittig B. Kleffe J, et al. BMC Bioinformatics. 2007 May 24;8 Suppl 5(Suppl 5):S7. doi: 10.1186/1471-2105-8-S5-S7. BMC Bioinformatics. 2007. PMID: 17570866 Free PMC article. - ECgene: genome-based EST clustering and gene modeling for alternative splicing.
Kim N, Shin S, Lee S. Kim N, et al. Genome Res. 2005 Apr;15(4):566-76. doi: 10.1101/gr.3030405. Genome Res. 2005. PMID: 15805497 Free PMC article.
References
- Adams M.D., Kelley,J.M., Gocayne,J.D., Dubnick,M., Polymeropoulos,M.H., Xiao,H., Merril,C.R., Wu,A., Olde,B., Moreno,R.F. et al. (1991) Complementary DNA sequencing: expressed sequence tags and human genome project. Science, 252, 1651–1656. - PubMed
- Venter J.C., Adams,M.D., Myers,E.W., Li,P.W., Mural,R.J., Sutton,G.G., Smith,H.O., Yandell,M., Evans,C.A., Holt,R.A., et al. (2001)The sequence of the human genome. Science, 291, 1304–1351. - PubMed
- Boguski M.S., Lowe,T.M. and Tolstoshev,C.M. (1993) dbEST–database for ‘expressed sequence tags’. Nature Genet., 4, 332–333. - PubMed
- Boguski M.S. (1995) The turning point in genome research. Trends Biochem. Sci., 20, 295–296. - PubMed
- Marra M.A., Hillier,L. and Waterston,R.H. (1998) Expressed sequence tags—ESTablishing bridges between genomes. Trends Genet., 14, 4–7. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials