Single primer amplification (SPA) of cDNA for microarray expression analysis - PubMed (original) (raw)
Comparative Study
Single primer amplification (SPA) of cDNA for microarray expression analysis
Lee Smith et al. Nucleic Acids Res. 2003.
Abstract
The potential of expression analysis using cDNA microarrays to address complex problems in a wide variety of biological contexts is now being realised. A limiting factor in such analyses is often the amount of RNA required, usually tens of micrograms. To address this problem researchers have turned to methods of improving detection sensitivity, either through increasing fluorescent signal output per mRNA molecule or increasing the amount of target available for labelling by use of an amplification procedure. We present a novel DNA-based method in which an oligonucleotide is incorporated into the 3' end of cDNA during second-strand cDNA synthesis. This sequence provides an annealing site for a single complementary heel primer that directs Taq DNA polymerase amplification of cDNA following multiple cycles of denaturation, annealing and extension. The utility of this technique for transcriptome-wide screening of relative expression levels was compared to two alternative methodologies for production of labelled cDNA target, namely incorporation of fluorescent nucleotides by reverse transcriptase or the Klenow fragment. Labelled targets from two distinct mouse tissues, adult liver and kidney, were compared by hybridisation to a set of cDNA microarrays containing 6500 mouse cDNA probes. Here we demonstrate, through a dilution series of cDNA derived from 10 micro g of total RNA, that it is possible to produce datasets comparable to those produced with unamplified targets with the equivalent of 30 ng of total RNA. The utility of this technique for microarray analysis in cases where sample is limited is discussed.
Figures
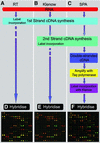
Figure 1
(A) A schematic overview of the three methods compared. In the RT experiments first-strand cDNA is fluorescently labelled through incorporation of Cy3-dCTP or Cy5-dCTP by reverse transcriptase. Samples are combined and hybridised to the microarray. (B) For the Klenow experiments cDNA is fluorescently labelled through random primed Klenow extension following first-strand cDNA synthesis. (C) In the SPA experiments double-stranded cDNA is entered for amplification and the resultant products labelled by random primed Klenow extension. (D–F) False colour overlay images of the same subsection of the array hybridised with cDNA derived from (D) 100 µg total RNA labelled with RT cDNA, (E) 20 µg of total RNA labelled with Klenow and (F) SPA cDNA equivalent to 0.031 µg total RNA.
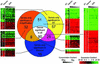
Figure 2
Reproducibility of outlier detection is demonstrated by comparison of genes selected as significant from each methodology [gene lists selected from individual slides using the methods described by Newton et al. (18) are combined if selected in all slides within each methodology to create the three segments of the Venn diagram]. The Venn diagram shows numbers of genes selected as significant in one, two or all three of the methodologies. Raster plots for the gene lists in each sector of the Venn diagram were created using single linkage clustering of each gene list in Cluster and Treeview (20). These plots demonstrate the variability of the ratio for a gene across each of the approaches used. Genes with relatively higher expression in kidney compared to liver are shown in red, while genes with relatively higher expression in liver compared to kidney are displayed in green. An expanded version of this figure listing the accession numbers of individual genes is available at
www.mgu.har.mrc.ac.uk/microarray/amplification
.
Similar articles
- An alternative method to amplify RNA without loss of signal conservation for expression analysis with a proteinase DNA microarray in the ArrayTube format.
Schüler S, Wenz I, Wiederanders B, Slickers P, Ehricht R. Schüler S, et al. BMC Genomics. 2006 Jun 12;7:144. doi: 10.1186/1471-2164-7-144. BMC Genomics. 2006. PMID: 16768788 Free PMC article. - Gene expression analysis using cDNA microarrays.
Karsten SL, Geshwind DH. Karsten SL, et al. Curr Protoc Neurosci. 2002 Nov;Chapter 4:Unit 4.28. doi: 10.1002/0471142301.ns0428s20. Curr Protoc Neurosci. 2002. PMID: 18428560 Review. - [Analysis of effectiveness of cDNA synthesis, induced using complementary primers and primers containing a noncomplementary base matrix].
D'iachenko LB, Chenchik AA, Khaspekov GL, Tatarenko AO, Bibilashvili RSh. D'iachenko LB, et al. Mol Biol (Mosk). 1994 Sep-Oct;28(5):1014-27. Mol Biol (Mosk). 1994. PMID: 7527482 Russian. - Oligonucleotide microarray analysis of aminoallyl-labeled cDNA targets from linear RNA amplification.
Kaposi-Novak P, Lee JS, Mikaelyan A, Patel V, Thorgeirsson SS. Kaposi-Novak P, et al. Biotechniques. 2004 Oct;37(4):580, 582-6, 588. doi: 10.2144/04374ST02. Biotechniques. 2004. PMID: 15517970 - [Polymerase chain reaction, cold probes and clinical diagnosis].
Haras D, Amoros JP. Haras D, et al. Sante. 1994 Jan-Feb;4(1):43-52. Sante. 1994. PMID: 7909267 Review. French.
Cited by
- Novel gene expression patterns along the proximo-distal axis of the mouse embryo before gastrulation.
Frankenberg S, Smith L, Greenfield A, Zernicka-Goetz M. Frankenberg S, et al. BMC Dev Biol. 2007 Feb 15;7:8. doi: 10.1186/1471-213X-7-8. BMC Dev Biol. 2007. PMID: 17302988 Free PMC article. - An efficient algorithm for the stochastic simulation of the hybridization of DNA to microarrays.
Arslan E, Laurenzi IJ. Arslan E, et al. BMC Bioinformatics. 2009 Dec 10;10:411. doi: 10.1186/1471-2105-10-411. BMC Bioinformatics. 2009. PMID: 20003312 Free PMC article. - Differentially expressed cDNAs at the early stage of banana ripening identified by suppression subtractive hybridization and cDNA microarray.
Xu BY, Su W, Liu JH, Wang JB, Jin ZQ. Xu BY, et al. Planta. 2007 Jul;226(2):529-39. doi: 10.1007/s00425-007-0502-6. Epub 2007 Mar 3. Planta. 2007. PMID: 17334781 - A robust method for the amplification of RNA in the sense orientation.
Marko NF, Frank B, Quackenbush J, Lee NH. Marko NF, et al. BMC Genomics. 2005 Mar 1;6:27. doi: 10.1186/1471-2164-6-27. BMC Genomics. 2005. PMID: 15740627 Free PMC article. - Options available for profiling small samples: a review of sample amplification technology when combined with microarray profiling.
Nygaard V, Hovig E. Nygaard V, et al. Nucleic Acids Res. 2006 Feb 9;34(3):996-1014. doi: 10.1093/nar/gkj499. Print 2006. Nucleic Acids Res. 2006. PMID: 16473852 Free PMC article. Review.
References
- Lockhart D. and Winzeler,E. (2000) Genomics, gene expression and DNA arrays. Nature, 405, 827–836. - PubMed
- Mills J.C., Roth,K.A., Cagan,R.L. and Gordon,J.I. (2001) DNA microarrays and beyond: completing the journey from tissue to cell. Nature Cell Biol., 3, E175–E178. - PubMed
- Schulze A. and Downward,J. (2001) Navigating gene expression using microarrays – a technology review. Nature Cell Biol., 3, E190–E195. - PubMed
- Stears R.L., Getts,R.C. and Gullans,S.R. (2000) A novel, sensitive detection system for high-density microarrays using dendrimer technology. Physiol. Genomics, 3, 93–99. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources