The maize genome contains a helitron insertion - PubMed (original) (raw)
The maize genome contains a helitron insertion
Shailesh K Lal et al. Plant Cell. 2003 Feb.
Abstract
The maize mutation sh2-7527 was isolated in a conventional maize breeding program in the 1970s. Although the mutant contains foreign sequences within the gene, the mutation is not attributable to an interchromosomal exchange or to a chromosomal inversion. Hence, the mutation was caused by an insertion. Sequences at the two Sh2 borders have not been scrambled or mutated, suggesting that the insertion is not caused by a catastrophic reshuffling of the maize genome. The insertion is large, at least 12 kb, and is highly repetitive in maize. As judged by hybridization, sorghum contains only one or a few copies of the element, whereas no hybridization was seen to the Arabidopsis genome. The insertion acts from a distance to alter the splicing of the sh2 pre-mRNA. Three distinct intron-bearing maize genes were found in the insertion. Of most significance, the insertion bears striking similarity to the recently described DNA helicase-bearing transposable elements termed HELITRONS: Like Helitrons, the inserted sequence of sh2-7527 is large, lacks terminal repeats, does not duplicate host sequences, and was inserted between a host dinucleotide AT. Like Helitrons, the maize element contains 5' TC and 3' CTRR termini as well as two short palindromic sequences near the 3' terminus that potentially can form a 20-bp hairpin. Although the maize element lacks sequence information for a DNA helicase, it does contain four exons with similarity to a plant DEAD box RNA helicase. A second Helitron insertion was found in the maize genomic database. These data strongly suggest an active Helitron in the present-day maize genome.
Figures
Figure 1.
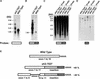
Insertion of sh2-7527. (A) RNA gel blot analysis of wild-type and sh2-7527 RNA. Total endosperm RNA, 20 days after pollination, was subjected to electrophoresis, blotted, and probed with a full-length Sh2 cDNA probe. MW, molecular mass. (B) The blot from (A) was stripped and probed with the PCR-derived, 1.2-kb, non-Sh2 portion of the sh2-7527 cDNA. (C) Genomic DNA from wild-type and sh2-7527 plants was digested with the indicated enzymes, electrophoresed, blotted, and hybridized with the PCR-derived, 1.2-kb, non-Sh2 portion of the sh2-7527 cDNA. (D) The blot from (C) was stripped and probed with a 3′ portion of the insertion of sh2-7527. (E) Scheme of wild-type and sh2-7527 cDNAs. The PCR-amplified sh2-7527 regions used to generate the probes are shown.
Figure 2.
Borders of the sh2-7527 Insertion. The sequences of the insertion and the flanking Sh2 intron 11 are shown in red and black letters, respectively. Conserved sequences at the termini of the element are underlined. Palindromic sequences capable of forming a hairpin are shown in blue letters.
Figure 3.
Scheme of sh2-7527. (A) The location of the sh2-7527 insertion within intron 11 of Sh2 is shown as a triangle. Exons are boxed. Exons of the ORF identified in the 3′ portion are listed from right to left. (B) Expanded version of the 5′ portion. Exons of Sh2 and the sh2-7527 insertion are boxed. Sh2-7527 insertion sequences are shown in bold. Exon P1 refers to a sequence with strong similarity to RNA helicase that is not found in the mature chimeric transcript. Exons with similarity to helicase and the sorting nexin are underlined.
Figure 4.
Spliced Alignment Results for the 5′ End of the sh2-7527 Insertion. The figure was produced with the MyGV viewer (Zhu and Brendel, 2002) for GeneSeqer (Usuka and Brendel, 2000; Usuka et al., 2000). Predicted exons are represented by boxes, with colors indicating various similarity scores (red, >0.9; pink, >0.8; cyan, >0.7; and gray, >0.6). vcdna+ indicates the sequenced transcript (cf. Figure 3B), helicase indicates Arabidopsis RNA helicase At1g00660, and sorting-nexin indicates Arabidopsis protein At5g06140. The other gene structures are spliced alignments with ESTs identified by their GenBank gi numbers followed by + or − to indicate the aligned strand. Vertical bars delineate introns and are of proportional length to their predicted splice site strengths. Spliced alignment to predict gene structure using the same tools is reviewed by Brendel and Zhu (2002).
Figure 5.
Alignment of the sh2-7527 Genomic Insertion Sequence with the Arabidopsis DEAD Box RNA Helicase (Aubourg et al., 1999). The alignment was produced by the program GeneSeqer (Usuka and Brendel, 2000). In the alignment, codons and amino acids representing conserved substitutions are marked +, and neutral substitutions are marked with dots between the amino acids. Predicted introns are not translated.
Figure 6.
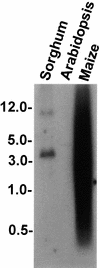
Presence of the sh2-7527 Insertion in Maize, Sorghum, and Arabidopsis. Genomic DNA from leaves was digested with EcoRI, electrophoresed, blotted, and hybridized with cDNA derived from the sh2-7527 insertion. Fragment sizes (kb) are shown at left.
Comment in
- A new twist on transposons: the maize genome harbors helitron insertion.
Eckardt NA. Eckardt NA. Plant Cell. 2003 Feb;15(2):293-5. doi: 10.1105/tpc.150210. Plant Cell. 2003. PMID: 12566572 Free PMC article. No abstract available.
Similar articles
- A novel class of Helitron-related transposable elements in maize contain portions of multiple pseudogenes.
Gupta S, Gallavotti A, Stryker GA, Schmidt RJ, Lal SK. Gupta S, et al. Plant Mol Biol. 2005 Jan;57(1):115-27. doi: 10.1007/s11103-004-6636-z. Plant Mol Biol. 2005. PMID: 15821872 - The polychromatic Helitron landscape of the maize genome.
Du C, Fefelova N, Caronna J, He L, Dooner HK. Du C, et al. Proc Natl Acad Sci U S A. 2009 Nov 24;106(47):19916-21. doi: 10.1073/pnas.0904742106. Epub 2009 Nov 19. Proc Natl Acad Sci U S A. 2009. PMID: 19926866 Free PMC article. - A new twist on transposons: the maize genome harbors helitron insertion.
Eckardt NA. Eckardt NA. Plant Cell. 2003 Feb;15(2):293-5. doi: 10.1105/tpc.150210. Plant Cell. 2003. PMID: 12566572 Free PMC article. No abstract available. - The splicing of maize transposable elements from pre-mRNA--a minireview.
Wessler SR. Wessler SR. Gene. 1989 Oct 15;82(1):127-33. doi: 10.1016/0378-1119(89)90037-1. Gene. 1989. PMID: 2555263 Review. - DNA methylation and activity of the maize Spm transposable element.
Fedoroff NV. Fedoroff NV. Curr Top Microbiol Immunol. 1995;197:143-64. doi: 10.1007/978-3-642-79145-1_10. Curr Top Microbiol Immunol. 1995. PMID: 7493489 Review. No abstract available.
Cited by
- Genetic dissection of intermated recombinant inbred lines using a new genetic map of maize.
Fu Y, Wen TJ, Ronin YI, Chen HD, Guo L, Mester DI, Yang Y, Lee M, Korol AB, Ashlock DA, Schnable PS. Fu Y, et al. Genetics. 2006 Nov;174(3):1671-83. doi: 10.1534/genetics.106.060376. Epub 2006 Sep 1. Genetics. 2006. PMID: 16951074 Free PMC article. - Differential pre-mRNA Splicing Alters the Transcript Diversity of Helitrons Between the Maize Inbred Lines.
Lynch BT, Patrick TL, Moreno JJ, Siebert AE, Klusman KM, Shodja DN, Hannah LC, Lal SK. Lynch BT, et al. G3 (Bethesda). 2015 Jun 12;5(8):1703-11. doi: 10.1534/g3.115.018630. G3 (Bethesda). 2015. PMID: 26070844 Free PMC article. - Nearly identical paralogs: implications for maize (Zea mays L.) genome evolution.
Emrich SJ, Li L, Wen TJ, Yandeau-Nelson MD, Fu Y, Guo L, Chou HH, Aluru S, Ashlock DA, Schnable PS. Emrich SJ, et al. Genetics. 2007 Jan;175(1):429-39. doi: 10.1534/genetics.106.064006. Epub 2006 Nov 16. Genetics. 2007. PMID: 17110490 Free PMC article. - Gene movement by Helitron transposons contributes to the haplotype variability of maize.
Lai J, Li Y, Messing J, Dooner HK. Lai J, et al. Proc Natl Acad Sci U S A. 2005 Jun 21;102(25):9068-73. doi: 10.1073/pnas.0502923102. Epub 2005 Jun 10. Proc Natl Acad Sci U S A. 2005. PMID: 15951422 Free PMC article. - Diverse DNA transposons in rotifers of the class Bdelloidea.
Arkhipova IR, Meselson M. Arkhipova IR, et al. Proc Natl Acad Sci U S A. 2005 Aug 16;102(33):11781-6. doi: 10.1073/pnas.0505333102. Epub 2005 Aug 4. Proc Natl Acad Sci U S A. 2005. PMID: 16081532 Free PMC article.
References
- Boeke, J., and Corces, V. (1989). Transcription and reverse transcription of retrotransposons. Annu. Rev. Microbiol. 43, 403–434. - PubMed
- Brendel, V., Carle-Urioste, J.C., and Walbot, V. (1998). Intron recognition in plants. In A Look beyond Transcription: Mechanisms Determining mRNA Stability and Translation in Plants, J. Bailey-Serres and D.R. Gallie, eds (Rockville, MD: American Society of Plant Physiologists), pp. 20–28.
- Brendel, V., and Zhu, W. (2002). Computational modeling of gene structure in Arabidopsis thaliana. Plant Mol. Biol. 48, 49–58. - PubMed
- Brink, R.A., and Burnham, C.R. (1929). Inheritance of semi sterility in maize. Am. Nat. 63, 301–316.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials