Early-replicating heterochromatin - PubMed (original) (raw)
Early-replicating heterochromatin
Soo-Mi Kim et al. Genes Dev. 2003.
Abstract
Euchromatin, which has an open structure and is frequently transcribed, tends to replicate in early S phase. Heterochromatin, which is more condensed and rarely transcribed, usually replicates in late S phase. Here, we report significant deviation from this correlation in the fission yeast, Schizosaccharomyces pombe. We found that heterochromatic centromeres and silent mating-type cassettes replicate in early S phase. Only heterochromatic telomeres replicate in late S phase. Research in other laboratories has shown that occasionally other organisms also replicate some of their heterochromatin in early S phase. Thus, late replication is not an obligatory feature of heterochromatin.
Figures
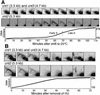
Figure 1
The central portions of the centromeres are replicated in early S phase. (A) The cdc10 temperature block and release procedure (Kim and Huberman 2001) was used to synchronize passage through S phase. This experiment is identical to the one shown in Figure 4 in Kim and Huberman (2001). The Southern membrane used in that experiment was stripped and rehybridized with probes specific for the cnt sequences. (B) The hydroxyurea (HU) block and release procedure (Kim and Huberman 2001) was used to synchronize passage through S phase. This experiment is identical to the one shown in Figure 7 in Kim and Huberman (2001). The Southern membrane used in that experiment was stripped and rehybridized with probes specific for the cnt sequences. Note that the cells in the zero-minute time point had been exposed to HU for 5 h at 25°C.
Figure 2
Structures of the major fission yeast mating-type locus configurations. These diagrams are based on nucleotide sequence information (GenBank accession nos. AL035065, AL356712, AL353012, and U57841) and on previous studies describing the derivation of h−S and h+N by recombination from h90 (Beach and Klar 1984). All three configurations are shown at the same scale. Restriction fragments studied for replication timing are indicated by horizontal boxes in the upper portions of the diagrams. The position of the hybridization probe used to identify each restriction fragment is shown as a thick black line immediately below the restriction fragment. The horizontally striped arrow in the CEN homology region represents dg sequences. The vertically striped arrow represents dh sequences, and the black arrowhead indicates an otr repeat that is frequently located between dh and dg sequences. See the text for additional description.
Figure 3
The mating-type region is replicated in early S phase. Locations of the studied restriction fragments are shown in Figure 2. (A) The cdc10 block and release procedure was used to synchronize the h−S strain. This experiment is identical to the one shown in Kim and Huberman (2001). The membrane was stripped and rehybridized with probes for the indicated restriction fragments. (B) The same procedure was used to synchronize the h+N strain. The experiment is the same as in Figure 5B in Kim and Huberman (2001). The membrane was stripped and rehybridized with probes for the indicated restriction fragments.
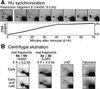
Figure 4
Early replication of the mating-type region. (A) The HU synchronization procedure was used with the h−S strain. This experiment is identical to the one shown in Figure 7 in Kim and Huberman (2001). The membrane was stripped and rehybridized with a probe for restriction fragment 2 (Fig. 2). (B) The centrifugal elutriation procedure was used with the h+N strain. This experiment is the same as in Figures 1 and 2 in Kim and Huberman (2001). The membrane was stripped and rehybridized with probes for the indicated regions. See the text and Materials and Methods for further details on the probes used.
Similar articles
- Role of heterochromatin in suppressing subtelomeric recombination in fission yeast.
Bisht KK, Arora S, Ahmed S, Singh J. Bisht KK, et al. Yeast. 2008 Aug;25(8):537-48. doi: 10.1002/yea.1603. Yeast. 2008. PMID: 18615848 - Fission yeast chromatin assembly factor 1 assists in the replication-coupled maintenance of heterochromatin.
Dohke K, Miyazaki S, Tanaka K, Urano T, Grewal SI, Murakami Y. Dohke K, et al. Genes Cells. 2008 Oct;13(10):1027-43. doi: 10.1111/j.1365-2443.2008.01225.x. Epub 2008 Aug 29. Genes Cells. 2008. PMID: 18761674 - A chromodomain protein, Chp1, is required for the establishment of heterochromatin in fission yeast.
Sadaie M, Iida T, Urano T, Nakayama J. Sadaie M, et al. EMBO J. 2004 Oct 1;23(19):3825-35. doi: 10.1038/sj.emboj.7600401. Epub 2004 Sep 16. EMBO J. 2004. PMID: 15372076 Free PMC article. - RNA interference and heterochromatin in the fission yeast Schizosaccharomyces pombe.
Martienssen RA, Zaratiegui M, Goto DB. Martienssen RA, et al. Trends Genet. 2005 Aug;21(8):450-6. doi: 10.1016/j.tig.2005.06.005. Trends Genet. 2005. PMID: 15979194 Review. - Silent chromatin at the middle and ends: lessons from yeasts.
Bühler M, Gasser SM. Bühler M, et al. EMBO J. 2009 Aug 5;28(15):2149-61. doi: 10.1038/emboj.2009.185. Epub 2009 Jul 23. EMBO J. 2009. PMID: 19629038 Free PMC article. Review.
Cited by
- Kinetochores coordinate pericentromeric cohesion and early DNA replication by Cdc7-Dbf4 kinase recruitment.
Natsume T, Müller CA, Katou Y, Retkute R, Gierliński M, Araki H, Blow JJ, Shirahige K, Nieduszynski CA, Tanaka TU. Natsume T, et al. Mol Cell. 2013 Jun 6;50(5):661-74. doi: 10.1016/j.molcel.2013.05.011. Mol Cell. 2013. PMID: 23746350 Free PMC article. - Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin.
Sullivan BA, Karpen GH. Sullivan BA, et al. Nat Struct Mol Biol. 2004 Nov;11(11):1076-83. doi: 10.1038/nsmb845. Epub 2004 Oct 10. Nat Struct Mol Biol. 2004. PMID: 15475964 Free PMC article. - Neocentromeres: a place for everything and everything in its place.
Scott KC, Sullivan BA. Scott KC, et al. Trends Genet. 2014 Feb;30(2):66-74. doi: 10.1016/j.tig.2013.11.003. Epub 2013 Dec 13. Trends Genet. 2014. PMID: 24342629 Free PMC article. Review. - Mechanisms Governing DDK Regulation of the Initiation of DNA Replication.
Larasati, Duncker BP. Larasati, et al. Genes (Basel). 2016 Dec 22;8(1):3. doi: 10.3390/genes8010003. Genes (Basel). 2016. PMID: 28025497 Free PMC article. Review. - Asynchronous replication timing of telomeres at opposite arms of mammalian chromosomes.
Zou Y, Gryaznov SM, Shay JW, Wright WE, Cornforth MN. Zou Y, et al. Proc Natl Acad Sci U S A. 2004 Aug 31;101(35):12928-33. doi: 10.1073/pnas.0404106101. Epub 2004 Aug 20. Proc Natl Acad Sci U S A. 2004. PMID: 15322275 Free PMC article.
References
- Allshire RC, Javerzat J-P, Redhead NJ, Cranston G. Position effect variegation at fission yeast centromeres. Cell. 1994;76:157–169. - PubMed
- Allshire RC, Nimmo ER, Ekwall K, Javerzat JP, Cranston G. Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes & Dev. 1995;9:218–233. - PubMed
- Baur JA, Zou Y, Shay JW, Wright WE. Telomere position effect in human cells. Science. 2001;292:2075–2077. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 49294/PHS HHS/United States
- P30 CA016056/CA/NCI NIH HHS/United States
- P30 CA16056-26/CA/NCI NIH HHS/United States
- CA095908/CA/NCI NIH HHS/United States
- R01 CA095908/CA/NCI NIH HHS/United States
- CA84302/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources