The ch-TOG/XMAP215 protein is essential for spindle pole organization in human somatic cells - PubMed (original) (raw)
The ch-TOG/XMAP215 protein is essential for spindle pole organization in human somatic cells
Fanni Gergely et al. Genes Dev. 2003.
Abstract
The ch-TOG/XMAP215 family of proteins bind directly to microtubules and appear to play an essential role in stabilizing spindle microtubules. These proteins stabilize microtubules mainly by influencing microtubule plus-end dynamics, yet, in vivo, they are all strongly concentrated at spindle poles, where the minus ends of the microtubules are concentrated. In Drosophila embryos, the centrosomal protein D-TACC is required to efficiently recruit ch-TOG/Msps to centrosomes. In humans, ch-TOG and the three known TACC proteins have been implicated in cancer, but their functions are unknown. Here we extensively depleted TACC3 and ch-TOG from HeLa cells using RNA interference. In TACC3-depleted cells, spindles are well organized, but microtubules are partially destabilized and ch-TOG is no longer concentrated on spindle microtubules. In ch-TOG-depleted cells, relatively robust spindles form, but the spindles are highly disorganized. Thus, in human somatic cells, ch-TOG appears to play a major role in organizing spindle poles, and a more minor role in stabilizing spindle microtubules that is, at least in part, mediated via an interaction with TACC3.
Figures
Figure 1
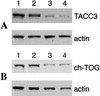
TACC3 and ch-TOG protein levels are reduced by siRNA treatment. A Western blot showing the levels of TACC3 (A) and ch-TOG (B) after 48 h of siRNA treatment. Actin is shown as a loading control. (A) Mock-depleted cells (lane 1), ch-TOG-depleted cells (lane 2), TACC3-depleted cells (lane 3), and TACC3- and ch-TOG-depleted cells (lane 4). (B) Mock-depleted cells (lane 1), TACC3-depleted cells (lane 2), ch-TOG-depleted cells (lane 3), and ch-TOG- and TACC3-depleted cells (lane 4).
Figure 2
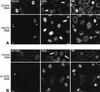
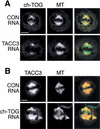
Immunofluorescence analysis of TACC3 and ch-TOG protein levels in siRNA-treated cells. (A) Mock-depleted (top panels) and TACC3-depleted (bottom panels) HeLa cells stained to reveal the distribution of TACC3, DNA, and microtubules. In this and all subsequent experiments, images were acquired from experimental and control cells that were treated and processed for immunofluorescence at the same time using identical settings on the confocal microscope. The TACC3 siRNA treatment has strongly reduced TACC3 levels in two mitotic cells (arrows), but substantial levels of TACC3 remain in one mitotic cell (arrowhead). (B) Mock-depleted (top panels) and ch-TOG-depleted (bottom panels) HeLa cells stained to reveal the distribution of ch-TOG, DNA, and microtubules. The ch-TOG siRNA treatment strongly reduced ch-TOG levels in most cells visible in this field, but small amounts of protein are still detectable on the centrosomes and spindles of some mitotic cells (arrowhead). Bar, 10 μm.
Figure 3
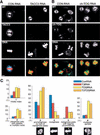
Mitotic defects in TACC3- and ch-TOG-depleted HeLa cells. (A,B) Mock-depleted (left panels) and either TACC3-depleted (A, right panels) or ch-TOG-depleted (B, right panels) cells were stained to reveal the distribution of microtubules (green in merged image), TACC3 or ch-TOG (blue in merged image), and DNA (red in merged image). Bar, 10 μm. (C) Quantitation of the defects observed in TACC3- and ch-TOG-depleted cells. In all graphs, the percentage of cells exhibiting a particular phenotype is indicated for mock-depleted (blue bars), TACC3-depleted (red bars), ch-TOG-depleted (yellow bars), or TACC3- and ch-TOG-depleted (orange bars) cells. (Graph 1) The mitotic index in the population of cells as determined by phospho-histone H3 staining. (Graph 2) The percentage of interphase cells exhibiting multiple or satellite nuclei. (Graph 3) The percentage of mitotic cells in a prometaphase-like state (with no discernible metaphase plate). (Graph 4) The percentage of mitotic cells with a normal metaphase plate. (Graph 5) The percentage of mitotic cells with a clear metaphase plate, but with at least one chromosome not aligned at the equator. (Graph 6) The percentage of mitotic cells with more than two spindle poles. Two-hundred-fifty cells were scored from each of four independent experiments for both the mitotic index and the multinucleated phenotype. Fifty metaphase cells from two independent experiments were scored for the prometaphase, metaphase, and lagging chromosome phenotypes. Thirty mitotic cells were scored from each of four independent experiments for the presence of multipolar spindles. Error bars represent the standard deviation.
Figure 4
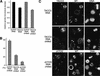
The depletion of TACC3 and ch-TOG partially destabilizes spindle microtubules. (A) A graph showing the relative spindle fluorescence intensity in nondepleted or TACC3-depleted mitotic cells (black bars), or in nondepleted or TACC3-depleted cells that have been chilled and allowed to recover for 25 min (hatched bars). Note that the spindle fluorescence of both sets of nondepleted cells has been normalized to 1. (B) A graph showing the percentage of cells that have properly aligned the majority of their chromosomes at a metaphase plate after 25 min of recovery from cold treatment. Error bars represent the standard deviation. (C, top panels) A field of cells treated with TACC3 siRNA. One mitotic cell (marked with *) has not been significantly depleted of TACC3, but two other mitotic cells (marked with arrowheads) have been significantly depleted. Fields such as these were used to compare the spindle microtubule density in the depleted and nondepleted cells (see Materials and Methods). Fields of TACC3 (middle panels) and ch-TOG (bottom panels) siRNA-treated cells that were chilled to depolymerize the microtubules and then allowed to recover for 25 min. In each panel, a nondepleted mitotic cell is marked with *, and depleted cells are marked with arrowheads. Bar, 10 μm.
Figure 5
The localization of ch-TOG is disrupted in TACC3-depleted cells. (A) The distribution of ch-TOG (left panels, red in merged image) and microtubules (middle panels, green in merged image) in mock-depleted (top panels) and TACC3-depleted (bottom panels) cells. Ch-TOG is detectable on centrosomes, but is not detectable on spindle microtubules in the TACC3-depleted cells. (B) The distribution of TACC3 (left panels, red in merged image) and microtubules (middle panels, green in merged image) in mock-depleted (top panels) and ch-TOG-depleted (bottom panels) cells. TACC3 remains concentrated on spindles in the ch-TOG-depleted cells. Bar, 10 μm.
Similar articles
- TACC3-ch-TOG interaction regulates spindle microtubule assembly by controlling centrosomal recruitment of γ-TuRC.
Rajeev R, Mukhopadhyay S, Bhagyanath S, Devu Priya MRS, Manna TK. Rajeev R, et al. Biosci Rep. 2023 Mar 29;43(3):BSR20221882. doi: 10.1042/BSR20221882. Biosci Rep. 2023. PMID: 36790370 Free PMC article. - Msps/XMAP215 interacts with the centrosomal protein D-TACC to regulate microtubule behaviour.
Lee MJ, Gergely F, Jeffers K, Peak-Chew SY, Raff JW. Lee MJ, et al. Nat Cell Biol. 2001 Jul;3(7):643-9. doi: 10.1038/35083033. Nat Cell Biol. 2001. PMID: 11433296 - MCAK-independent functions of ch-Tog/XMAP215 in microtubule plus-end dynamics.
Barr AR, Gergely F. Barr AR, et al. Mol Cell Biol. 2008 Dec;28(23):7199-211. doi: 10.1128/MCB.01040-08. Epub 2008 Sep 22. Mol Cell Biol. 2008. PMID: 18809577 Free PMC article. - The role of TACC3 in mitotic spindle organization.
Ding ZM, Huang CJ, Jiao XF, Wu D, Huo LJ. Ding ZM, et al. Cytoskeleton (Hoboken). 2017 Oct;74(10):369-378. doi: 10.1002/cm.21388. Epub 2017 Aug 23. Cytoskeleton (Hoboken). 2017. PMID: 28745816 Review. - The role of TOG domains in microtubule plus end dynamics.
Slep KC. Slep KC. Biochem Soc Trans. 2009 Oct;37(Pt 5):1002-6. doi: 10.1042/BST0371002. Biochem Soc Trans. 2009. PMID: 19754440 Review.
Cited by
- Kinesin-6 Klp9 orchestrates spindle elongation by regulating microtubule sliding and growth.
Krüger LK, Gélin M, Ji L, Kikuti C, Houdusse A, Théry M, Blanchoin L, Tran PT. Krüger LK, et al. Elife. 2021 Jun 3;10:e67489. doi: 10.7554/eLife.67489. Elife. 2021. PMID: 34080538 Free PMC article. - ZYG-9ch-TOG promotes the stability of acentrosomal poles via regulation of spindle microtubules in C. elegans oocyte meiosis.
Cavin-Meza G, Mullen TJ, Czajkowski ER, Wolff ID, Divekar NS, Finkle JD, Wignall SM. Cavin-Meza G, et al. PLoS Genet. 2022 Nov 30;18(11):e1010489. doi: 10.1371/journal.pgen.1010489. eCollection 2022 Nov. PLoS Genet. 2022. PMID: 36449516 Free PMC article. - CKAP5 stabilizes CENP-E at kinetochores by regulating microtubule-chromosome attachments.
Lakshmi RB, Nayak P, Raz L, Sarkar A, Saroha A, Kumari P, Nair VM, Kombarakkaran DP, Sajana S, M G S, Agasti SS, Paul R, Ben-David U, Manna TK. Lakshmi RB, et al. EMBO Rep. 2024 Apr;25(4):1909-1935. doi: 10.1038/s44319-024-00106-9. Epub 2024 Feb 29. EMBO Rep. 2024. PMID: 38424231 Free PMC article. - TACC3 is required for the proper mitosis of sclerotome mesenchymal cells during formation of the axial skeleton.
Yao R, Natsume Y, Noda T. Yao R, et al. Cancer Sci. 2007 Apr;98(4):555-62. doi: 10.1111/j.1349-7006.2007.00433.x. Epub 2007 Mar 14. Cancer Sci. 2007. PMID: 17359303 Free PMC article. - TACC3 is essential for EGF-mediated EMT in cervical cancer.
Ha GH, Kim JL, Breuer EK. Ha GH, et al. PLoS One. 2013 Aug 1;8(8):e70353. doi: 10.1371/journal.pone.0070353. Print 2013. PLoS One. 2013. PMID: 23936413 Free PMC article.
References
- Brinkley BR, Goepfert TM. Supernumerary centrosomes and cancer: Boveri's hypothesis resurrected. Cell Motil Cytoskeleton. 1998;41:281–288. - PubMed
- Charrasse S, Schroeder M, Gauthier-Rouviere C, Ango F, Cassimeris L, Gard DL, Larroque C. The TOGp protein is a new human microtubule-associated protein homologous to the Xenopus XMAP215. J Cell Sci. 1998;111:1371–1383. - PubMed
- Ciciarello M, Mangiacasale R, Casenghi M, Zaira Limongi M, D'Angelo M, Soddu S, Lavia P, Cundari E. p53 displacement from centrosomes and p53-mediated G1 arrest following transient inhibition of the mitotic spindle. J Biol Chem. 2001;276:19205–19213. - PubMed
- Cullen CF, Ohkura H. Msps protein is localized to acentrosomal poles to ensure bipolarity of Drosophila meiotic spindles. Nat Cell Biol. 2001;3:637–642. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases