Tracking an embryonic L1 retrotransposition event - PubMed (original) (raw)
Tracking an embryonic L1 retrotransposition event
Eline T Luning Prak et al. Proc Natl Acad Sci U S A. 2003.
Abstract
Long interspersed nuclear elements 1 (L1) are active retrotransposons that reside in many species, including humans and rodents. L1 elements produce an RNA intermediate that is reverse transcribed to DNA and inserted in a new genomic location. We have tagged an active human L1 element (L1(RP)) with a gene encoding enhanced GFP (EGFP). Expression of GFP occurs only if L1-EGFP has undergone a cycle of transcription, reverse transcription, and integration into a transcriptionally permissive genomic region. We show here that L1-EGFP can undergo retrotransposition in vivo and produce fluorescence in mouse testis. The retrotransposition event characterized here has occurred at a very early stage in the development of an L1-EGFP transgenic founder mouse.
Figures
Figure 1
Retrotransposition of L1-EGFP. (a) Schematic of the L1-EGFP transgene and its retrotransposition. L1 transcription is driven by the mouse RNA pol II promoter in addition to the L1 5′ UTR (21). The EGFP gene is in the antisense orientation relative to L1. EGFP (green) is situated in the 3′ UTR (hatched) of L1 and is interrupted by the mouse γ-globin intron. The intron is in the same transcriptional orientation as L1 (18). Therefore, when the L1 sense-strand transcript is processed, the γ-globin intron is spliced out. The EGFP gene is driven by the human CMV MIE promoter (pCMV-MIE) and has an HSV thymidine kinase polyadenylation sequence (tkpA). pCMV-MIE, EGFP, and tkpA are all antisense relative to L1RP. At the very 3′ end of the L1-EGFP transgene is the SV40 late polyadenylation sequence (SV40pA) derived from the pCEP4 cloning vector (Invitrogen). 5′ truncation of the L1-EGFP insertion is depicted with a jagged line. Arrows depict the locations of the geno5 (left) and geno3 (right) genotyping primers used in the PCR assay shown in b (not drawn to scale). (b) The geno5 and geno3 primers flank the intron in EGFP and give rise to two products, a 1.5-kb amplicon (corresponding to the intron-containing transgene) and an ≈600-bp amplicon that lacks the 909-bp intron (corresponding to the insertion). Shown are the genotyping results on tail DNA from five offspring of founder 57 (lanes 1–5). dw, distilled water; neg, genomic DNA from the tail of a transgene negative mouse; tg, L1-EGFP transgene; XIV, 100-bp ladder with bright bands at 500 bp, 1,000 bp, and 2.6 kb (top band; Roche).
Figure 2
Nucleotide sequence of the L1-EGFP insertion flanks. The genomic sequences flanking the L1-EGFP insertion are lowercase. L1-EGFP sequences adjoining the flanks are given in uppercase. The 5′ flanking sequence is shown in the top portion of the figure, followed by the 3′ flanking sequence in the bottom. Each flank is numbered separately. The complete L1-EGFP sequence (between the 5′ and the 3′ flanks) is not shown. The discontinuity corresponding to the L1-EGFP sequence is denoted by three asterisks at the end of the 5′ flank. Target site duplications (TSD) are in bold. The thymine at position 422, just downstream of the 5′ TSD, is not present in the L1RP sequence at that position. A nucleotide
blast
search of the mouse genome database using sequences flanking the L1-EGFP insertion suggests that the insertion is on Mus musculus chromosome 4 (
).
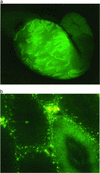
Figure 3
EGFP fluorescence in the seminiferous tubules of Fo57. (a) Stereofluorescent image (×20), testis (fresh tissue) from Fo57. (b) Fluorescent light image (×200, frozen section from Fo57 testis. GFP fluorescence (dark green) is restricted to the seminiferous tubule in the lower right. Interstitial areas are autofluorescent (yellow–green speckled pattern).
Figure 4
Tissue DNA PCR surveys in Fo57. Genomic DNA from Fo57 tissues (liver, intestine, cerebrum, and lung) was amplified with geno5 and geno3 primers. A short extension time was used to favor the smaller amplicon (corresponding to the L1-EGFP insertion) relative to the larger amplicon (corresponding to the transgene). MW, 100-bp molecular weight ladder; dw, water; 2205, tail DNA from a line 57 mouse that has the transgene (present in one to five copies) and the single copy insertion.
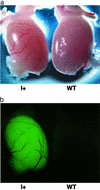
Figure 5
EGFP fluorescence in the testes of mice inheriting the L1-EGFP insertion. (a) Transmitted light image, testis from an F1 mouse with only the insertion (I+, son of Fo57), and a WT littermate. (b) Stereofluorescent light image, same testis pair as in a.
Similar articles
- Retrotransposon RNA expression and evidence for retrotransposition events in human oocytes.
Georgiou I, Noutsopoulos D, Dimitriadou E, Markopoulos G, Apergi A, Lazaros L, Vaxevanoglou T, Pantos K, Syrrou M, Tzavaras T. Georgiou I, et al. Hum Mol Genet. 2009 Apr 1;18(7):1221-8. doi: 10.1093/hmg/ddp022. Epub 2009 Jan 15. Hum Mol Genet. 2009. PMID: 19147684 - Retrotransposon expression and incorporation of cloned human and mouse retroelements in human spermatozoa.
Lazaros L, Kitsou C, Kostoulas C, Bellou S, Hatzi E, Ladias P, Stefos T, Markoula S, Galani V, Vartholomatos G, Tzavaras T, Georgiou I. Lazaros L, et al. Fertil Steril. 2017 Mar;107(3):821-830. doi: 10.1016/j.fertnstert.2016.12.027. Epub 2017 Jan 27. Fertil Steril. 2017. PMID: 28139237 - Evaluation of LINE-1 mobility in neuroblastoma cells by in vitro retrotransposition reporter assay: FACS analysis can detect only the tip of the iceberg of the inserted L1 elements.
Del Re B, Marcantonio P, Capri M, Giorgi G. Del Re B, et al. Exp Cell Res. 2010 Dec 10;316(20):3358-67. doi: 10.1016/j.yexcr.2010.06.024. Epub 2010 Jul 8. Exp Cell Res. 2010. PMID: 20620136 - L1 retrotransposons and somatic mosaicism in the brain.
Richardson SR, Morell S, Faulkner GJ. Richardson SR, et al. Annu Rev Genet. 2014;48:1-27. doi: 10.1146/annurev-genet-120213-092412. Epub 2014 Jul 14. Annu Rev Genet. 2014. PMID: 25036377 Review.
Cited by
- Repeat-induced gene silencing of L1 transgenes is correlated with differential promoter methylation.
Rosser JM, An W. Rosser JM, et al. Gene. 2010 May 15;456(1-2):15-23. doi: 10.1016/j.gene.2010.02.005. Epub 2010 Feb 16. Gene. 2010. PMID: 20167267 Free PMC article. - Gamma radiation increases endonuclease-dependent L1 retrotransposition in a cultured cell assay.
Farkash EA, Kao GD, Horman SR, Prak ET. Farkash EA, et al. Nucleic Acids Res. 2006 Feb 28;34(4):1196-204. doi: 10.1093/nar/gkj522. Print 2006. Nucleic Acids Res. 2006. PMID: 16507671 Free PMC article. - Cell divisions are required for L1 retrotransposition.
Shi X, Seluanov A, Gorbunova V. Shi X, et al. Mol Cell Biol. 2007 Feb;27(4):1264-70. doi: 10.1128/MCB.01888-06. Epub 2006 Dec 4. Mol Cell Biol. 2007. PMID: 17145770 Free PMC article. - LINE-1 retrotransposons: mediators of somatic variation in neuronal genomes?
Singer T, McConnell MJ, Marchetto MC, Coufal NG, Gage FH. Singer T, et al. Trends Neurosci. 2010 Aug;33(8):345-54. doi: 10.1016/j.tins.2010.04.001. Epub 2010 May 12. Trends Neurosci. 2010. PMID: 20471112 Free PMC article. - The Role of Somatic L1 Retrotransposition in Human Cancers.
Scott EC, Devine SE. Scott EC, et al. Viruses. 2017 May 31;9(6):131. doi: 10.3390/v9060131. Viruses. 2017. PMID: 28561751 Free PMC article. Review.
References
- Kazazian H H, Moran J V. Nat Genet. 1998;19:19–23. - PubMed
- Ostertag E, Kazazian H H., Jr Annu Rev Genet. 2001;35:501–538. - PubMed
- Sassaman D M, Dombroski B A, Moran J V, Kimberland M L, Naas T P, DeBerardinis R J, Gabriel A, Swergold G D, Kazazian H H., Jr Nat Genet. 1997;16:37–43. - PubMed
- Lander E S, Linton L M, Birren B, Nusbaum C, Zody M C, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Nature. 2001;409:860–921. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources