Mapping the binding interface between human eukaryotic initiation factors 1A and 5B: a new interaction between old partners - PubMed (original) (raw)
Mapping the binding interface between human eukaryotic initiation factors 1A and 5B: a new interaction between old partners
Assen Marintchev et al. Proc Natl Acad Sci U S A. 2003.
Abstract
The translation initiation factors (IFs) IF1/eIF1A and IF2e/IF5B have been conserved throughout all kingdoms. Although the central roles of the bacterial factors IF1 and IF2 were established long ago, the importance of their eukaryotic homologs, eukaryotic IFs (eIFs) eIF1A and eIF5B, has only recently become evident. The translation machinery in eukaryotes is more complex and accordingly, eIF1A and eIF5B seem to have acquired a number of new functions while also retaining many of the roles of bacterial IF1 and IF2. IF1 and IF2 have been shown to interact on the ribosome but no binding has been detected for the free factors. In contrast, yeast eIF1A and eIF5B have been reported to interact in the absence of ribosomes. Here, we have identified the binding interface between human eIF1A and the C-terminal domain of eIF5B by using solution NMR. That interaction interface involves the C termini of the two proteins, which are not present in bacterial IF1 and IF2. The interaction is, therefore, unique to eukaryotes. A structural model for the interaction of eIF1A and eIF5B in the context of the ribosome is presented. We propose that eIF1A and eIF5B simultaneously interact at two sites that are >50 A apart: through their C termini as reported here, and through an interface previously identified in bacterial IF1 and IF2. The binding between the C termini of eIF1A and eIF5B has implications for eukaryote-specific mechanisms of recruitment and release of translation IFs from the ribosome.
Figures
Figure 1
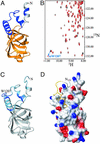
Binding of human eIF5B-CTD to 15N-/13C-ILV-methyl-labeled human eIF1A. (A) Segment of the 15N-HSQC spectrum of eIF1A, in the presence (red) and absence (black) of unlabeled eIF5B-CTD. The segment contains all peaks, whose positions were affected. The initial and final positions of the peaks corresponding to the NH groups of residues 139–143 are connected by arrows. Gly-134 (the most N-terminal residue to show any effect) is also shown. (B) 13C-HSQC spectrum of 13C-ILV-methyl-labeled eIF1A, in the presence (red) and absence (black) of unlabeled eIF5B-CTD. The peaks corresponding to the δ-methyl groups of Ile-140 and Ile-143 are indicated as in A. (C) Mapping the effects of eIF5B-CTD binding on the structure of eIF1A (8). Residues affected by eIF5B-CTD binding are colored from yellow (small chemical-shift changes) to orange (big changes). The side chains of Ile-140 and Ile-143 are shown.
Figure 2
Binding of human eIF1A to 15N-labeled human eIF5B-CTD. (A) Homology-based model of the structure of human eIF5B-CTD. The model was created as described in Materials and Methods, using the x-ray structure of the archaeal IF2/eIF5B homolog (12) as a template. Residues belonging to the helix connecting eIF5B-CTD to domain III are colored in gray; the eIF5B-CTD is colored in orange; and segments present in the archaeal and eukaryotic IF2/eIF5B homologs, but not in bacteria (12), are colored in dark blue. NMR data indicated that the first seven residues were not helical (as in the model) but unstructured, and this is reflected in C and D. (B) 15N-HSQC spectrum of protein G-tagged eIF5B-CTD, in the presence (red) and absence (black) of an unlabeled C-terminal eIF1A peptide FDDIGDDDEDIDDI. As an example, the initial and final positions of the side-chain NH peak of Trp-1207 (which had the greatest chemical-shift change) are connected by an arrow. (C) Mapping the effects of eIF1A binding on the structure of eIF5B-CTD. Residues affected by eIF1A binding are colored from light blue (small chemical-shift changes) to dark blue (big changes). The side chain of Trp-1207 is shown. Big changes (Δσ > 0.1 ppm) were observed for residues 1188, 1191, 1197, 1203, 1207, and 1216. Smaller changes (Δσ < 0.1 ppm) were observed for residues 1076–1082, 1088–1090, 1092, 1118, 1186, 1189, 1192, 1194, 1195, 1198–1202, 1204, 1209–1215, and 1220. (D) Interaction interface between human eIF1A and eIF5B-CTD. eIF5B-CTD, in the same orientation as in C, is in surface representation and colored by electrostatic potential. Positive charges are in blue and negative charges are in red. The C-terminal 14 residues of eIF1A are displayed but only the last five residues were used in the docking, because the rest of the C terminus of eIF1A remains unstructured in the complex. The coloring of eIF1A is as in Fig. 1_C_. The side chains of amino acids 138–143 are displayed.
Figure 3
Model for the interactions of IF1/eIF1A and IF2/eIF5B on the ribosome. (A) Conservation of IF1/eIF1A and IF2/eIF5B throughout evolution. The structures of bacterial IF1 (7), human eIF1A (8), and the archaeal IF2/eIF5B homolog (12) are shown. The individual domains of eIF5B are painted from yellow (domain I) to dark orange (domain IV) and the connecting segments between the domains are in gray. Segments in eIF1A and eIF5B painted in blue, as well as the N-terminal segment of eIF1A (painted in gray), are not present in bacteria. The arrows indicate the interactions between IF1 and IF2 (Upper) and eIF1A and eIF5B (Lower). (B) Surface representation of the proposed orientation of IF1/eIF1A and IF2/eIF5B on the ribosome. The structure of eIF5B (painted in orange) was docked on the x-ray structure of the 70S bacterial ribosome (28), by orienting the G domain (domain I) toward the L7/L12 stalk on the large ribosomal subunit (not shown); the CTD (domain IV) toward the acceptor end of the P-site tRNA (red); and domain II toward the A-site of the small subunit (painted in gray). The structure of human eIF1A (blue) was aligned to the structure of IF1, and this alignment was used to replace IF1 with eIF1A in the IF1 cocrystal structure with the 30S ribosomal subunit (4). The location of the positively charged N-terminal segment of eIF1A (not shown) is not known. The unstructured C-terminal tail of eIF1A was extended and directed along the A-site groove in the ribosome toward the CTD of eIF5B. The C terminus of eIF1A was able to reach several angstroms beyond its binding site on eIF5B-CTD.
Similar articles
- Remarkable conservation of translation initiation factors: IF1/eIF1A and IF2/eIF5B are universally distributed phylogenetic markers.
Sørensen HP, Hedegaard J, Sperling-Petersen HU, Mortensen KK. Sørensen HP, et al. IUBMB Life. 2001 May;51(5):321-7. doi: 10.1080/152165401317190842. IUBMB Life. 2001. PMID: 11699879 - Physical and functional interaction between the eukaryotic orthologs of prokaryotic translation initiation factors IF1 and IF2.
Choi SK, Olsen DS, Roll-Mecak A, Martung A, Remo KL, Burley SK, Hinnebusch AG, Dever TE. Choi SK, et al. Mol Cell Biol. 2000 Oct;20(19):7183-91. doi: 10.1128/MCB.20.19.7183-7191.2000. Mol Cell Biol. 2000. PMID: 10982835 Free PMC article. - Domains of eIF1A that mediate binding to eIF2, eIF3 and eIF5B and promote ternary complex recruitment in vivo.
Olsen DS, Savner EM, Mathew A, Zhang F, Krishnamoorthy T, Phan L, Hinnebusch AG. Olsen DS, et al. EMBO J. 2003 Jan 15;22(2):193-204. doi: 10.1093/emboj/cdg030. EMBO J. 2003. PMID: 12514125 Free PMC article. - Structural insights on the translation initiation complex: ghosts of a universal initiation complex.
Allen GS, Frank J. Allen GS, et al. Mol Microbiol. 2007 Feb;63(4):941-50. doi: 10.1111/j.1365-2958.2006.05574.x. Mol Microbiol. 2007. PMID: 17238926 Review. - Structural insights into the evolution of late steps of translation initiation in the three domains of life.
Kazan R, Bourgeois G, Lazennec-Schurdevin C, Coureux PD, Mechulam Y, Schmitt E. Kazan R, et al. Biochimie. 2024 Feb;217:31-41. doi: 10.1016/j.biochi.2023.02.002. Epub 2023 Feb 9. Biochimie. 2024. PMID: 36773835 Review.
Cited by
- Human eIF5 and eIF1A Compete for Binding to eIF5B.
Lin KY, Nag N, Pestova TV, Marintchev A. Lin KY, et al. Biochemistry. 2018 Oct 9;57(40):5910-5920. doi: 10.1021/acs.biochem.8b00839. Epub 2018 Sep 26. Biochemistry. 2018. PMID: 30211544 Free PMC article. - Poliovirus switches to an eIF2-independent mode of translation during infection.
White JP, Reineke LC, Lloyd RE. White JP, et al. J Virol. 2011 Sep;85(17):8884-93. doi: 10.1128/JVI.00792-11. Epub 2011 Jun 22. J Virol. 2011. PMID: 21697471 Free PMC article. - Recent Advances in Archaeal Translation Initiation.
Schmitt E, Coureux PD, Kazan R, Bourgeois G, Lazennec-Schurdevin C, Mechulam Y. Schmitt E, et al. Front Microbiol. 2020 Sep 18;11:584152. doi: 10.3389/fmicb.2020.584152. eCollection 2020. Front Microbiol. 2020. PMID: 33072057 Free PMC article. Review. - Sulfolobus solfataricus translation initiation factor 1 stimulates translation initiation complex formation.
Hasenöhrl D, Benelli D, Barbazza A, Londei P, Bläsi U. Hasenöhrl D, et al. RNA. 2006 Apr;12(4):674-82. doi: 10.1261/rna.2289306. Epub 2006 Mar 3. RNA. 2006. PMID: 16517972 Free PMC article. - Crystallization and preliminary X-ray crystallographic analysis of eIF5BΔN and the eIF5BΔN-eIF1AΔN complex.
Zheng A, Yamamoto R, Sokabe M, Tanaka I, Yao M. Zheng A, et al. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2011 Jun 1;67(Pt 6):730-3. doi: 10.1107/S1744309111015910. Epub 2011 May 26. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2011. PMID: 21636924 Free PMC article.
References
- Hershey J W B, Merrick W C. In: Translational Control of Gene Expression. Sonenberg N, Hershey J W B, Mathews M B, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 2000. pp. 33–88.
- Roll-Mecak A, Shin B S, Dever T E, Burley S K. Trends Biochem Sci. 2001;26:705–709. - PubMed
- Carter A P, Clemons W M, Jr, Brodersen D E, Morgan-Warren R J, Hartsch T, Wimberly B T, Ramakrishnan V. Science. 2001;291:498–501. - PubMed
- Moazed D, Samaha R R, Gualerzi C, Noller H F. J Mol Biol. 1995;248:207–210. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases