STAT5-induced Id-1 transcription involves recruitment of HDAC1 and deacetylation of C/EBPbeta - PubMed (original) (raw)
STAT5-induced Id-1 transcription involves recruitment of HDAC1 and deacetylation of C/EBPbeta
Min Xu et al. EMBO J. 2003.
Abstract
Transcriptional activation is associated commonly with recruitment of histone acetylases, while repression involves histone deacetylases (HDACs). Here, we provide evidence to suggest that STAT5 activates gene expression by recruiting HDAC. The interleukin-3 (IL-3)-dependent expression of the Id-1 gene, encoding a helix-loop-helix (HLH) transcriptional inhibitor, is activated by both C/EBPbeta and STAT5 transcription factors bound to its pro-B-cell enhancer (PBE), but is inhibited by HDAC inhibitors in Ba/F3 cells. STAT5 interacts with HDAC1 in the PBE region, resulting in deacetylation of histones, as well as C/EBPbeta, whose acetylation diminishes its DNA-binding activity. Consistently, expression of an acetylation-resistant mutant of C/EBPbeta results in IL-3-independent expression of the Id-1 gene. Thus, we propose a novel mechanism by which STAT5 mediates the deacetylation of C/EBPbeta, allowing transcriptional activation.
Figures
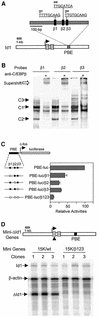
Fig. 1. The C/EBPβ-binding site is required for Id-1 expression. (A) Three C/EBPβ-binding sites are diagrammed as filled boxes, and their sequences shown on top. Mutations are indicated with lower case letters. The position of PBE (gray box) relative to Id-1 exons (hatched boxes) is illustrated. (B) EMSA with each of the three C/EBPβ-binding sites as probes. A 70 bp probe containing the β1, β2 or β3 site was incubated with Ba/F3 nuclear extracts with or without antibodies against C/EBPβ. The binding reactions were analyzed on a 6% poly acrylamide gel in Tris–glycine buffer. C/EBPβ-containing complexes are labeled as C1–C3. Supershifted complexes are marked with an open arrow. Additional bands are non-specific complexes found in all cell types. (C) Mutational analysis in transient transfection assays using Ba/F3 cells. Constructs containing the 460 bp PBE placed upstream of a luciferase reporter gene driven by a c-fos minimal promoter are diagrammed. Filled and open boxes represent wild-type and mutant C/EBPβ sites, respectively. Luciferase activities were normalized against the β-galactosidase activity. The data are presented as activities relative to that of the PBE-luc construct and are averages of at least three independent experiments with standard deviations. (D) Mutational analysis in stable Ba/F3 cell lines. The ΔId-1 gene is diagrammed as in (A). Three independent cell lines stably transfected with the indicated constructs were analyzed using RPAs, and transcripts were detected with the probes as labeled.
Fig. 2. Identities of the C1–C3 complexes. (A) EMSA was performed with nuclear extracts from NIH-3T3 cells transfected with the indicated constructs. ‘C’ stands for extracts transfected with an empty vector, pcDNA3. The positions of the indicated homo- and heterodimers of LAP and LIP are labeled on the left, while C1–C3 complexes in Ba/F3 cells are marked on the right. Supershifted complexes with antibodies against C/EBPβ are indicated by an open arrow. Nuclear extracts used in lanes 4 and 5 were analyzed using a western blot with antibodies against the C-terminus of C/EBPβ. The doublets represent the 29 and 31 kDa LAP. (B) ChIP assays with antibodies against C/EBPβ and a negative control, Skp2. A fragment containing all three C/EBP-binding sites was amplified. (C) Transient co-transfection assay of Ba/F3 cells with the indicated constructs; normalized luciferase activities are shown relative to that of PBE-luc.
Fig. 3. STAT5 mediates IL-3-dependent Id1 expression. (A) Northern and western blots (NB and WB) for the levels of Id-1 mRNA and phospho-STAT5 protein in Ba/F3 cells cultured without IL-3 for the indicated times. The levels of GAPDH mRNA and C/EBPβ protein were used as internal controls. Twenty percent of WEHI-3 conditioned medium was used as a source of IL-3 in all experiments. (B) Northern blots for Id-1 and GAPDH mRNA levels in Ba/F3 cells: lane 1, continuously cultured in IL-3; lane 2, deprived of IL-3 for 5 h; lane 3, as lane 2 plus treatment with CHX at 50 µg/ml for the last 80 min; lane 4, re-stimulated with IL-3 for 60 min after a 4 h starvation; and lane 5, as lane 4 plus treatment with CHX at 50 µg/ml for the last 80 min.
Fig. 4. STAT5 binds to PBE. (A) Identification of STAT5-binding sites. Three potential sites shown in triangles, named S1–S3, were found within PBE (thin line). The three C/EBPβ-binding sites are marked as in Figure 1. EMSA was performed using end-labeled 30 bp oligonucleotides containing the S1 site. STAT5 binding and supershifted complexes are indicated by filled and open arrows. Unlabeled 30 bp oligonucleotides containing the S1, S2 or S3 site, plus a known STAT5 site, were used as competitors at the indicated molar excess to the probe. (B) ChIP assays with anti-STAT5a antibodies. Fragments containing the S1 site (S1), S2 plus S3 sites (S23) and C/EBP sites (C/EBP), as diagrammed in (A), were amplified using DNA templates before (input) or after immunoprecipitation (IP) with anti-STAT5 and control antibodies. (C) Effect of deletion of STAT sites on PBE activity. Left: Ba/F3 cells were transfected with the indicated constructs along with the CMV-LacZ construct. Luciferase activities are normalized with that of β-galactosidase and are shown as averages of activities relative to that of PBE-luc obtained from at least three independent experiments. Right: C/EBP-dependent activity is the ratio of relative activities between enhancers with wild-type and mutant C/EBP sites (open and closed ovals).
Fig. 5. Deacetylase activities are necessary for PBE-mediated transcription of the Id-1 gene. (A) ChIP assays of the Id-1 locus depicted as in Figure 1. PCR fragments corresponding to various regions in the locus are shown as short black bars and labeled A–C. Each fragment was amplified from immunoprecipitates of Ba/F3 cells cultured in medium plus or minus IL-3. Fragment B was also amplified using DNA preparations prior to IP as input controls. Data shown are representative of three independent experiments with the indicated antibodies. (B) Northern blot analyses of Id-1 and GAPDH levels in 4 h IL-3-deprived Ba/F3 cells, which were re-stimulated with (lanes 5–7) or without (lanes 2–4) IL-3 for 1 h. TSA (lane 3, 4, 6 and 7) was added 1 h before and during re-stimulation at the indicated concentrations. Lane 1 contains RNA from Ba/F3 cells continuously cultured with IL-3. The percentage Id-1 re-stimulation (lanes 5–7) was calculated by comparing Id-1 levels normalized with levels of GAPDH and shown in the bar graph. (C) Northern blots to measure the levels of Id-2 and GAPDH mRNA. Treatments in lanes 1–6 were identical to those in lanes 2–7 of (B). (D) Transient transfection assays for expression of the indicated reporter genes under the following conditions: (1) continuous culture in IL-3; (2) IL-3 deprivation for 16 h; (3) IL-3 deprivation for 10 h followed by 6 h re-stimulation; (4) the same as (3) plus TSA treatment 1 h before and during IL-3 re-stimulation. SEAP activities accumulated in media during the last 6 h of the treatment were normalized against β-galactosidase activities. Data are presented as activities relative to SEAP activities under condition 1.
Fig. 6. Interaction between STAT5 and HDAC1. (A) Total cell extracts derived from Ba/F3 cells cultured with or without (+ or –) IL-3 were precipitated with the indicated antibodies. Immunoprecipitates and inputs were analyzed using western blots with antibodies against HDAC1 or STAT5a. (B) 293T cells were transfected with the indicated constructs, and nuclear extracts were analyzed after IP with antibodies against HDAC1 and western blotting with anti-STAT5 antibodies. Inputs were analyzed using western blots with the indicated antibodies. (C) ChIP assay was performed using antibodies against HDAC1 and Ba/F3 cells cultured with or without IL-3. PCR products for regions A and B were as described in Figure 5A.
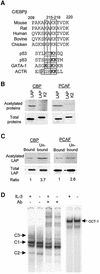
Fig. 7. Acetylation of C/EBPβ. (A) Sequence alignment of a potential acetylation site in C/EBPβ and other proteins as indicated. The three conserved lysines in the motif are boxed. Acetylation motifs are underlined, with acetylated lysine in bold. (B) In vitro acetylation. Wild-type and mutant LAP were incubated with the indicated acetyltransferases and [14C]acetyl-CoA, and labeled proteins were analyzed by SDS–PAGE followed by autoradiography. Input proteins (3%) were analyzed by western blots. The mutant LAP protein contains an HA tag, thus migrating more slowly than LAP. (C) Acetylation inhibits the DNA-binding activity of LAP–LAP dimers. Pull-down assays were performed with agarose beads conjugated with C/EBPβ-binding sites and in vitro acetylated LAP proteins. Equal portions of the bound and unbound fractions were analyzed using SDS–PAGE and quantified with a phosphoimager. The amounts of proteins in these fractions were also analyzed by western blots and quantified with a LumiImager. The amount of acetylated LAP was normalized against that of total LAP in each fraction. The ratio between the proportion of acetylated LAP in the unbound and bound fractions is shown below. (D) EMSA using nuclear extracts from Ba/F3 cells cultured with (+) or without (–) IL-3. Different forms of C/EBPβ dimers were identified by supershifting with anti-C/EBPβ antibodies. Oct-1-binding activities were used as controls for the nuclear extracts.
Fig. 8. An acetylation mutant of C/EBPβ enables IL-3-independent expression of Id-1. (A) In vivo acetylation assay. Immunoprecipitates of an anti-acetyl-lysine antibody (KAc) and a control antibody (anti-E47) from nuclear extracts of Ba/F3 (B) or PD31 (P) cells were analyzed using western blots with a monoclonal antibody against the C-terminus of C/EBPβ. Inputs contain 2% of nuclear extracts used for IP. (B) Real-time PCR assays for Id-1 expression in Ba/F3 cells expressing HA-tagged wild-type or K2 mutant C/EBPβ. The experimental procedure is as outlined. Data shown are averages of three real-time PCR assays with standard deviations, and expressed as levels relative to that at time zero. Protein expression at each time point was analyzed using western blots with an antibody against C/EBPβ. The endogenous (endo) and exogenous (exo) LAP proteins are indicated by arrows. The level of TFIIH serves as a loading control. (C) Acetylation of wild-type and K2 mutant protein. Ba/F3 cells infected with retrovectors expressing HA-tagged wild-type and K2 mutant were used to immunoprecipitate the proteins with an anti-HA antibody. The immunoprecipitates were probed with anti-acetyl-lysine and anti-HA antibodies.
Similar articles
- Glucocorticoid receptor (GR)-associated SMRT binding to C/EBPbeta TAD and Nrf2 Neh4/5: role of SMRT recruited to GR in GSTA2 gene repression.
Ki SH, Cho IJ, Choi DW, Kim SG. Ki SH, et al. Mol Cell Biol. 2005 May;25(10):4150-65. doi: 10.1128/MCB.25.10.4150-4165.2005. Mol Cell Biol. 2005. PMID: 15870285 Free PMC article. - Characterization of BCE-1, a transcriptional enhancer regulated by prolactin and extracellular matrix and modulated by the state of histone acetylation.
Myers CA, Schmidhauser C, Mellentin-Michelotti J, Fragoso G, Roskelley CD, Casperson G, Mossi R, Pujuguet P, Hager G, Bissell MJ. Myers CA, et al. Mol Cell Biol. 1998 Apr;18(4):2184-95. doi: 10.1128/MCB.18.4.2184. Mol Cell Biol. 1998. PMID: 9528790 Free PMC article. - Regulating the Regulators: The Role of Histone Deacetylase 1 (HDAC1) in Erythropoiesis.
Kim MY, Yan B, Huang S, Qiu Y. Kim MY, et al. Int J Mol Sci. 2020 Nov 11;21(22):8460. doi: 10.3390/ijms21228460. Int J Mol Sci. 2020. PMID: 33187090 Free PMC article. Review.
Cited by
- Signal transducer and activator of transcription STAT5 is recruited to c-Myc super-enhancer.
Pinz S, Unser S, Rascle A. Pinz S, et al. BMC Mol Biol. 2016 Apr 14;17:10. doi: 10.1186/s12867-016-0063-y. BMC Mol Biol. 2016. PMID: 27074708 Free PMC article. - Roles and regulation of stat family transcription factors in human breast cancer.
Clevenger CV. Clevenger CV. Am J Pathol. 2004 Nov;165(5):1449-60. doi: 10.1016/S0002-9440(10)63403-7. Am J Pathol. 2004. PMID: 15509516 Free PMC article. Review. - HDAC stimulates gene expression through BRD4 availability in response to IFN and in interferonopathies.
Marié IJ, Chang HM, Levy DE. Marié IJ, et al. J Exp Med. 2018 Dec 3;215(12):3194-3212. doi: 10.1084/jem.20180520. Epub 2018 Nov 21. J Exp Med. 2018. PMID: 30463877 Free PMC article. - The biological functions of the versatile transcription factors STAT3 and STAT5 and new strategies for their targeted inhibition.
Desrivières S, Kunz C, Barash I, Vafaizadeh V, Borghouts C, Groner B. Desrivières S, et al. J Mammary Gland Biol Neoplasia. 2006 Jan;11(1):75-87. doi: 10.1007/s10911-006-9014-4. J Mammary Gland Biol Neoplasia. 2006. PMID: 16947086 Review. - CCAAT/enhancer-binding protein beta: its role in breast cancer and associations with receptor tyrosine kinases.
Zahnow CA. Zahnow CA. Expert Rev Mol Med. 2009 Apr 8;11:e12. doi: 10.1017/S1462399409001033. Expert Rev Mol Med. 2009. PMID: 19351437 Free PMC article. Review.
References
- Boyes J., Byfield,P., Nakatani,Y. and Ogryzko,V. (1998) Regulation of activity of the transcription factor GATA-1 by acetylation. Nature, 396, 594–598. - PubMed
- Bromberg J. and Darnell,J.E.,Jr (2000) The role of STATs in transcriptional control and their impact on cellular function. Oncogene, 19, 2468–2473. - PubMed
- Chen H., Lin,R.J., Xie,W., Wilpitz,D. and Evans,R.M. (1999) Regulation of hormone-induced histone hyperacetylation and gene activation via acetylation of an acetylase. Cell, 98, 675–686. - PubMed
- Chen H., Tini,M. and Evans,R.M. (2001) HATs on and beyond chromatin. Curr. Opin. Cell Biol., 13, 218–224. - PubMed
- Chen L., Fischle,W., Verdin,E. and Greene,W.C. (2001) Duration of nuclear NF-κB action regulated by reversible acetylation. Science, 293, 1653–1657. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI033597/AI/NIAID NIH HHS/United States
- CA77553/CA/NCI NIH HHS/United States
- R21 AI033597/AI/NIAID NIH HHS/United States
- AI33597/AI/NIAID NIH HHS/United States
- R01 CA077553/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous