Extensive linkage disequilibrium, a common 16.7-kilobase deletion, and evidence of balancing selection in the human protocadherin alpha cluster - PubMed (original) (raw)
Comparative Study
doi: 10.1086/368060. Epub 2003 Feb 7.
Affiliations
- PMID: 12577201
- PMCID: PMC1180238
- DOI: 10.1086/368060
Comparative Study
Extensive linkage disequilibrium, a common 16.7-kilobase deletion, and evidence of balancing selection in the human protocadherin alpha cluster
James P Noonan et al. Am J Hum Genet. 2003 Mar.
Abstract
Regions of extensive linkage disequilibrium (LD) appear to be a common feature of the human genome. However, the mechanisms that maintain these regions are unknown. In an effort to understand whether gene density contributes to LD, we determined the degree of promoter sequence variation in a large tandem-arrayed gene family, the human protocadherin alpha cluster, on chromosome 5. These genes are expressed at synaptic junctions in the developing brain and the adult brain and may be involved in the determination of synaptic complexity. We sequenced the promoters of all 13 alpha protocadherin genes in 96 European Americans and identified polymorphisms in the promoters alpha 1, alpha 3, alpha 4, alpha 5, alpha 7, alpha 9, alpha 11, and alpha 13. In these promoters, 11 common SNPs are in extensive LD, forming two 48-kb haplotypes of equal frequency, in this population, that extend from the alpha1 through alpha 7 genes. We sequenced these promoters in East Asians and African Americans, and we estimated haplotype frequencies and calculated LD statistics for all three populations. Our results indicate that, although extensive LD is an ancient feature of the alpha cluster, it has eroded over time. SNPs 3' of alpha 7 are involved in ancestral recombination events in all populations, and overall alpha-cluster LD is reduced in African Americans. We obtained significant positive values for Tajima's D test for all alpha promoter SNPs in Europeans (D=3.03) and East Asians (D=2.64), indicating an excess of intermediate-frequency variants, which is a signature of balancing selection. We also discovered a 16.7-kb deletion that truncates the alpha 8 gene and completely removes the alpha 9 and alpha 10 genes. This deletion appears in unaffected individuals from multiple populations, suggesting that a reduction in protocadherin gene number is not obviously deleterious.
Figures
Figure 1
Structure and organization of the human protocadherin α genes. A, The human protocadherin α cluster. Each black box represents a single short-form exon; smaller black boxes represent promoter sequences (not to scale). Positions of common promoter polymorphisms and the two major haplotypes that they comprise are shown. SNPs are numbered relative to the translation start site of each gene. Exons of the α constant region are positioned at the 3′ end of the α cluster as indicated (white boxes). αC1 and αC2 are more similar to each other than to the other members of the α cluster. Exon sizes and distributions are approximately to scale (except for constant-region exons). B, Protocadherin promoter structure. Each exon has a core promoter element (black) upstream of the translation start site. This core promoter is highly conserved among paralogs and orthologs. Nucleotides in boldface are almost completely conserved in all mouse and human α, β, and γ protocadherin-cluster promoters. Gray shading indicates regions conserved between specific mouse and human orthologous promoters.
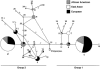
Figure 2
MS network for predicted α-cluster haplotypes. The size of each node is proportional to the haplotype frequency in the sample. Colors within each node represent the relative distribution of each haplotype among the three populations in the present study. Branch lengths represent one nucleotide substitution, except as otherwise labeled. Dashed lines represent alternative relationships among haplotypes. For clarity, in some cases, branches are not drawn to scale.
Figure 3
The α8–α10 deletion. A, The deletion interval. The region from α7 to α11 is shown, with known promoter polymorphisms positioned as indicated. Intergenic regions are not shown to scale. B, Structure of the α8–α10 deletion junction. Three nucleotides of unknown origin link position 1231 of α8 to a site 764 bases 3′ of the α10 stop codon.
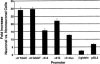
Figure 4
Fold increase in α3 and α9 promoter strength on neuronal differentiation of mouse P19 cells by use of retinoic acid. Results shown are the average of three independent experiments.
Similar articles
- Identification and characterization of coding single-nucleotide polymorphisms within human protocadherin-alpha and -beta gene clusters.
Miki R, Hattori K, Taguchi Y, Tada MN, Isosaka T, Hidaka Y, Hirabayashi T, Hashimoto R, Fukuzako H, Yagi T. Miki R, et al. Gene. 2005 Apr 11;349:1-14. doi: 10.1016/j.gene.2004.11.044. Gene. 2005. PMID: 15777644 - Similarity in recombination rate and linkage disequilibrium at CYP2C and CYP2D cytochrome P450 gene regions among Europeans indicates signs of selection and no advantage of using tagSNPs in population isolates.
Pimenoff VN, Laval G, Comas D, Palo JU, Gut I, Cann H, Excoffier L, Sajantila A. Pimenoff VN, et al. Pharmacogenet Genomics. 2012 Dec;22(12):846-57. doi: 10.1097/FPC.0b013e32835a3a6d. Pharmacogenet Genomics. 2012. PMID: 23089684 - Sequence variation and linkage disequilibrium in the GABA transporter-1 gene (SLC6A1) in five populations: implications for pharmacogenetic research.
Hirunsatit R, Ilomäki R, Malison R, Räsänen P, Ilomäki E, Kranzler HR, Kosten T, Sughondhabirom A, Thavichachart N, Tangwongchai S, Listman J, Mutirangura A, Gelernter J, Lappalainen J. Hirunsatit R, et al. BMC Genet. 2007 Oct 17;8:71. doi: 10.1186/1471-2156-8-71. BMC Genet. 2007. PMID: 17941974 Free PMC article. - Patterns of genetic variation in the hypertension candidate gene GRK4: ethnic variation and haplotype structure.
Lohmueller KE, Wong LJ, Mauney MM, Jiang L, Felder RA, Jose PA, Williams SM. Lohmueller KE, et al. Ann Hum Genet. 2006 Jan;70(Pt 1):27-41. doi: 10.1111/j.1529-8817.2005.00197.x. Ann Hum Genet. 2006. PMID: 16441255 - Nucleotide diversity and haplotype structure of the human angiotensinogen gene in two populations.
Nakajima T, Jorde LB, Ishigami T, Umemura S, Emi M, Lalouel JM, Inoue I. Nakajima T, et al. Am J Hum Genet. 2002 Jan;70(1):108-23. doi: 10.1086/338454. Epub 2001 Nov 30. Am J Hum Genet. 2002. PMID: 11731937 Free PMC article.
Cited by
- Characterization of X-linked SNP genotypic variation in globally distributed human populations.
Casto AM, Li JZ, Absher D, Myers R, Ramachandran S, Feldman MW. Casto AM, et al. Genome Biol. 2010 Jan 28;11(1):R10. doi: 10.1186/gb-2010-11-1-r10. Genome Biol. 2010. PMID: 20109212 Free PMC article. - The role of clustered protocadherins in neurodevelopment and neuropsychiatric diseases.
Flaherty E, Maniatis T. Flaherty E, et al. Curr Opin Genet Dev. 2020 Dec;65:144-150. doi: 10.1016/j.gde.2020.05.041. Epub 2020 Jul 14. Curr Opin Genet Dev. 2020. PMID: 32679536 Free PMC article. Review. - Kilquist syndrome: A novel syndromic hearing loss disorder caused by homozygous deletion of SLC12A2.
Macnamara EF, Koehler AE, D'Souza P, Estwick T, Lee P, Vezina G; Undiagnosed Diseases Network; Fauni H, Braddock SR, Torti E, Holt JM, Sharma P, Malicdan MCV, Tifft CJ. Macnamara EF, et al. Hum Mutat. 2019 May;40(5):532-538. doi: 10.1002/humu.23722. Epub 2019 Mar 12. Hum Mutat. 2019. PMID: 30740830 Free PMC article. - Molecular codes for neuronal individuality and cell assembly in the brain.
Yagi T. Yagi T. Front Mol Neurosci. 2012 Apr 12;5:45. doi: 10.3389/fnmol.2012.00045. eCollection 2012. Front Mol Neurosci. 2012. PMID: 22518100 Free PMC article. - In silico analysis of 2085 clones from a normalized rat vestibular periphery 3' cDNA library.
Roche JP, Wackym PA, Cioffi JA, Kwitek AE, Erbe CB, Popper P. Roche JP, et al. Audiol Neurootol. 2005 Nov-Dec;10(6):310-22. doi: 10.1159/000087348. Epub 2005 Aug 5. Audiol Neurootol. 2005. PMID: 16103642 Free PMC article.
References
Electronic-Database Information
- Arlequin, http://lgb.unige.ch/arlequin/
- dbSNP Home Page, http://www.ncbi.nlm.nih.gov/SNP/ (for ss5607025–ss5607061)
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for protocadherin α, β, and γ subclusters [accession numbers NG_000016, NG_000017, and NG_000012, respectively] and BAC DNA [accession number AC020968])
- KCL, Institute of Psychiatry, Section of Genetic Epidemiology and Biostatistics, http://www.iop.kcl.ac.uk/IoP/Departments/PsychMed/GEpiBSt/software.stm (for 2LD)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for the human protocadherin α subcluster [MIM 604966])
References
- Angst BD, Marcozzi C, Magee AI (2001) The cadherin superfamily: diversity in form and function. J Cell Sci 114:629–641 - PubMed
- Bain G, Yao M, Huettner JE, Finley MFA, Gottlieb DI (1998) Neuronlike cells derived in culture from P19 embryonal carcinoma and embryonic stem cells. In: Banker G, Goslin K (eds) Culturing nerve cells, 2nd ed. MIT Press, Cambridge, pp 189–212
- Beasley A, Myers RM, Cox DR, Lazzeroni LC (1999) Statistical refinement of primer design parameters. In: Innis MA, Gelfand DH, Sninsky JJ (eds) PCR applications. Academic Press, New York, pp 55–72
- Bowcock AM, Ruiz-Linares A, Tomfohrde J, Minch E, Kidd JR, Cavalli-Sforza LL (1994) High resolution of human evolutionary trees with polymorphic microsatellites. Nature 368:455–457 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials