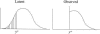A tobit variance-component method for linkage analysis of censored trait data - PubMed (original) (raw)
A tobit variance-component method for linkage analysis of censored trait data
Michael P Epstein et al. Am J Hum Genet. 2003 Mar.
Abstract
Variance-component (VC) methods are flexible and powerful procedures for the mapping of genes that influence quantitative traits. However, traditional VC methods make the critical assumption that the quantitative-trait data within a family either follow or can be transformed to follow a multivariate normal distribution. Violation of the multivariate normality assumption can occur if trait data are censored at some threshold value. Trait censoring can arise in a variety of ways, including assay limitation or confounding due to medication. Valid linkage analyses of censored data require the development of a modified VC method that directly models the censoring event. Here, we present such a model, which we call the "tobit VC method." Using simulation studies, we compare and contrast the performance of the traditional and tobit VC methods for linkage analysis of censored trait data. For the simulation settings that we considered, our results suggest that (1) analyses of censored data by using the traditional VC method lead to severe bias in parameter estimates and a modest increase in false-positive linkage findings, (2) analyses with the tobit VC method lead to unbiased parameter estimates and type I error rates that reflect nominal levels, and (3) the tobit VC method has a modest increase in linkage power as compared with the traditional VC method. We also apply the tobit VC method to censored data from the Finland-United States Investigation of Non-Insulin-Dependent Diabetes Mellitus Genetics study and provide two examples in which the tobit VC method yields noticeably different results as compared with the traditional method.
Figures
Figure 1
Example of censoring for a normally distributed trait. Latent trait values truly less than y* are observed at y*.
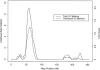
Figure 2
VC analyses of HDLR on chromosome 2. For the tobit method, the maximum likelihood-ratio statistic is 9.1 (maximum LOD score 1.98), at 58 cM; for the traditional method, the maximum likelihood-ratio statistic is 5.7 (maximum LOD score 1.24), at 60 cM.
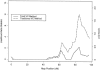
Figure 3
VC analyses of TG on chromosome 20. For the tobit method, the maximum likelihood-ratio statistic is 2.2 (maximum LOD score 0.48), at 85 cM; for the traditional method, the maximum likelihood-ratio statistic is 8.0 (maximum LOD score 1.74), at 84 cM.
Similar articles
- Quantitative trait linkage analysis using Gaussian copulas.
Li M, Boehnke M, Abecasis GR, Song PX. Li M, et al. Genetics. 2006 Aug;173(4):2317-27. doi: 10.1534/genetics.105.054650. Epub 2006 Jun 4. Genetics. 2006. PMID: 16751671 Free PMC article. - Semiparametric variance-component models for linkage and association analyses of censored trait data.
Diao G, Lin DY. Diao G, et al. Genet Epidemiol. 2006 Nov;30(7):570-81. doi: 10.1002/gepi.20168. Genet Epidemiol. 2006. PMID: 16858699 - Bayesian multilocus association mapping on ordinal and censored traits and its application to the analysis of genetic variation among Oryza sativa L. germplasms.
Iwata H, Ebana K, Fukuoka S, Jannink JL, Hayashi T. Iwata H, et al. Theor Appl Genet. 2009 Mar;118(5):865-80. doi: 10.1007/s00122-008-0945-6. Epub 2009 Jan 9. Theor Appl Genet. 2009. PMID: 19132337 - Genetic linkage methods for quantitative traits.
Amos CI, de Andrade M. Amos CI, et al. Stat Methods Med Res. 2001 Feb;10(1):3-25. doi: 10.1177/096228020101000102. Stat Methods Med Res. 2001. PMID: 11329691 Review. - Linkage mapping of quantitative trait loci in humans: an overview.
Ghosh S, Reich T, Majumder PP. Ghosh S, et al. Ann Hum Genet. 2002 Nov;66(Pt 5-6):431-8. doi: 10.1017/S0003480002001240. Ann Hum Genet. 2002. PMID: 12485475 Review.
Cited by
- Evaluation of a bayesian model integration-based method for censored data.
Hou L, Wang K, Bartlett CW. Hou L, et al. Hum Hered. 2012;74(1):1-11. doi: 10.1159/000342707. Epub 2012 Sep 26. Hum Hered. 2012. PMID: 23018141 Free PMC article. - A Variance-Component Framework for Pedigree Analysis of Continuous and Categorical Outcomes.
Epstein MP, Hunter JE, Allen EG, Sherman SL, Lin X, Boehnke M. Epstein MP, et al. Stat Biosci. 2009 Nov;1(2):181-198. doi: 10.1007/s12561-009-9010-5. Stat Biosci. 2009. PMID: 20436936 Free PMC article. - The effects of age on inflammatory and coagulation-fibrinolysis response in patients hospitalized for pneumonia.
Kale S, Yende S, Kong L, Perkins A, Kellum JA, Newman AB, Vallejo AN, Angus DC; GenIMS Investigators. Kale S, et al. PLoS One. 2010 Nov 4;5(11):e13852. doi: 10.1371/journal.pone.0013852. PLoS One. 2010. PMID: 21085465 Free PMC article. - Understanding the potential role of statins in pneumonia and sepsis.
Yende S, Milbrandt EB, Kellum JA, Kong L, Delude RL, Weissfeld LA, Angus DC. Yende S, et al. Crit Care Med. 2011 Aug;39(8):1871-8. doi: 10.1097/CCM.0b013e31821b8290. Crit Care Med. 2011. PMID: 21516038 Free PMC article. - Understanding the inflammatory cytokine response in pneumonia and sepsis: results of the Genetic and Inflammatory Markers of Sepsis (GenIMS) Study.
Kellum JA, Kong L, Fink MP, Weissfeld LA, Yealy DM, Pinsky MR, Fine J, Krichevsky A, Delude RL, Angus DC; GenIMS Investigators. Kellum JA, et al. Arch Intern Med. 2007 Aug 13-27;167(15):1655-63. doi: 10.1001/archinte.167.15.1655. Arch Intern Med. 2007. PMID: 17698689 Free PMC article.
References
- Agatston AS, Janowitz WR, Hildner FJ, Zusmer NR, Viamonte M Jr, Detrano R (1990) Quantification of coronary artery calcium using ultrafast computer tomography. J Am Coll Cardiol 15:827–832 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous